Project
DANIO-CODE
Navigation
Downloads
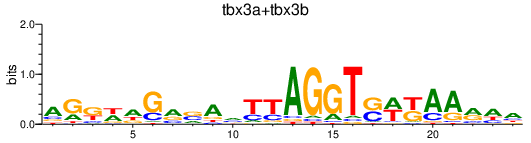
Results for tbx3a+tbx3b
Z-value: 0.70
Transcription factors associated with tbx3a+tbx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx3a
|
ENSDARG00000002216 | T-box transcription factor 3a |
|
tbx3b
|
ENSDARG00000061509 | T-box transcription factor 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx3a | dr10_dc_chr5_-_71539823_71539910 | -0.69 | 3.0e-03 | Click! |
| tbx3b | dr10_dc_chr5_+_26312951_26312966 | -0.60 | 1.4e-02 | Click! |
Activity profile of tbx3a+tbx3b motif
Sorted Z-values of tbx3a+tbx3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx3a+tbx3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_18664720 | 2.11 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr18_+_17545564 | 1.85 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr14_-_14353451 | 1.52 |
ENSDART00000170355
ENSDART00000159888 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr13_+_33331767 | 1.47 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr3_+_37648476 | 1.31 |
ENSDART00000151506
|
si:dkey-260c8.8
|
si:dkey-260c8.8 |
| chr2_-_39026651 | 1.30 |
ENSDART00000114085
|
si:ch211-119o8.6
|
si:ch211-119o8.6 |
| chr7_-_32598812 | 1.22 |
|
|
|
| chr10_+_15066791 | 1.21 |
ENSDART00000140084
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr3_+_37648665 | 1.18 |
ENSDART00000151506
|
si:dkey-260c8.8
|
si:dkey-260c8.8 |
| chr17_-_53241446 | 1.14 |
|
|
|
| chr8_-_49227232 | 1.13 |
|
|
|
| chr13_+_8508786 | 1.10 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr18_-_21759723 | 1.09 |
ENSDART00000079253
|
pskh1
|
protein serine kinase H1 |
| chr7_-_21571924 | 1.05 |
ENSDART00000166446
|
BX005336.1
|
ENSDARG00000102693 |
| chr19_-_18664670 | 1.02 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr23_-_27318392 | 1.02 |
|
|
|
| chr18_-_21759831 | 0.98 |
ENSDART00000079253
|
pskh1
|
protein serine kinase H1 |
| chr22_-_25015224 | 0.92 |
ENSDART00000015512
|
polr3f
|
polymerase (RNA) III (DNA directed) polypeptide F |
| chr3_-_44466637 | 0.88 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr25_+_32419008 | 0.88 |
|
|
|
| chr7_-_21572054 | 0.87 |
ENSDART00000166446
|
BX005336.1
|
ENSDARG00000102693 |
| chr15_-_5913264 | 0.82 |
ENSDART00000155156
ENSDART00000155971 |
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr2_-_21780380 | 0.80 |
ENSDART00000144587
|
plcd1b
|
phospholipase C, delta 1b |
| chr18_-_50439653 | 0.78 |
ENSDART00000151038
|
CU862078.1
|
ENSDARG00000096262 |
| chr21_+_25651637 | 0.76 |
ENSDART00000125709
|
si:dkey-17e16.17
|
si:dkey-17e16.17 |
| chr21_-_34938261 | 0.68 |
ENSDART00000023838
|
lipia
|
lipase, member Ia |
| chr23_-_43396787 | 0.67 |
|
|
|
| chr13_+_849563 | 0.66 |
|
|
|
| chr25_-_35094522 | 0.65 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr18_-_40923017 | 0.64 |
ENSDART00000098878
|
polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr18_-_7138350 | 0.63 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr1_+_52686620 | 0.62 |
ENSDART00000176087
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr17_-_53241366 | 0.62 |
ENSDART00000171082
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr23_-_27318468 | 0.60 |
|
|
|
| chr5_-_19987344 | 0.59 |
ENSDART00000144232
|
ficd
|
FIC domain containing |
| chr6_-_32099929 | 0.59 |
ENSDART00000150934
|
alg6
|
asparagine-linked glycosylation 6 (alpha-1,3,-glucosyltransferase) |
| chr5_+_2429397 | 0.58 |
|
|
|
| chr1_+_52686770 | 0.56 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr9_-_45104857 | 0.54 |
ENSDART00000131252
|
zgc:66484
|
zgc:66484 |
| chr14_-_14353487 | 0.54 |
ENSDART00000172241
|
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr8_+_7815256 | 0.53 |
ENSDART00000165575
|
cxxc1a
|
CXXC finger protein 1a |
| KN150380v1_-_10405 | 0.50 |
ENSDART00000168535
|
CABZ01048453.1
|
ENSDARG00000104434 |
| chr4_-_5766814 | 0.48 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr10_+_15066970 | 0.48 |
ENSDART00000140084
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr5_+_29203411 | 0.48 |
ENSDART00000134900
|
usf1l
|
upstream transcription factor 1, like |
| chr16_+_32230379 | 0.47 |
ENSDART00000084009
|
zufsp
|
zinc finger with UFM1-specific peptidase domain |
| chr17_-_43725091 | 0.46 |
|
|
|
| chr15_-_24007305 | 0.44 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr18_-_7138171 | 0.44 |
ENSDART00000149122
|
cfap161
|
cilia and flagella associated protein 161 |
| chr9_-_29510914 | 0.43 |
ENSDART00000122602
|
pth2r
|
parathyroid hormone 2 receptor |
| chr10_+_15066654 | 0.43 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr16_+_32230439 | 0.42 |
ENSDART00000084009
|
zufsp
|
zinc finger with UFM1-specific peptidase domain |
| chr18_-_7138309 | 0.42 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr2_+_21527785 | 0.41 |
ENSDART00000136498
|
si:dkey-29d8.3
|
si:dkey-29d8.3 |
| chr14_+_3409236 | 0.40 |
ENSDART00000158416
|
fnta
|
farnesyltransferase, CAAX box, alpha |
| chr3_-_26211047 | 0.40 |
ENSDART00000087196
|
zgc:153240
|
zgc:153240 |
| chr15_-_23588004 | 0.40 |
ENSDART00000078396
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr13_+_30033024 | 0.39 |
ENSDART00000040926
|
eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr15_-_5912917 | 0.37 |
|
|
|
| chr15_-_1800095 | 0.35 |
ENSDART00000149980
|
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr16_-_47366405 | 0.34 |
ENSDART00000169697
|
mios
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr4_-_18786577 | 0.34 |
ENSDART00000141187
|
def6c
|
differentially expressed in FDCP 6c homolog |
| chr4_+_15007453 | 0.31 |
ENSDART00000146156
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr12_-_26400022 | 0.30 |
ENSDART00000153361
|
si:dkey-287g12.6
|
si:dkey-287g12.6 |
| chr24_-_38758093 | 0.29 |
|
|
|
| chr25_+_3521763 | 0.29 |
ENSDART00000160017
|
CABZ01058261.1
|
ENSDARG00000099194 |
| chr11_-_205744 | 0.29 |
ENSDART00000172775
|
tegt
|
testis enhanced gene transcript (BAX inhibitor 1) |
| chr8_+_15213743 | 0.25 |
ENSDART00000063717
|
ENSDARG00000043402
|
ENSDARG00000043402 |
| chr18_-_7138414 | 0.24 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr22_+_39098205 | 0.24 |
ENSDART00000002826
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr7_+_56171081 | 0.23 |
ENSDART00000073596
ENSDART00000123273 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr1_+_24960066 | 0.20 |
ENSDART00000054235
|
plrg1
|
pleiotropic regulator 1 |
| chr18_-_40923158 | 0.19 |
ENSDART00000098878
|
polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr16_-_14697143 | 0.19 |
ENSDART00000012479
|
dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr15_-_24007488 | 0.18 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr1_-_2305324 | 0.17 |
ENSDART00000152555
|
ggact.3
|
gamma-glutamylamine cyclotransferase, tandem duplicate 3 |
| chr24_-_9146200 | 0.17 |
ENSDART00000153817
|
CU459174.1
|
ENSDARG00000097146 |
| chr2_+_42155813 | 0.17 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr15_-_1800139 | 0.16 |
ENSDART00000093074
|
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr23_-_19905270 | 0.15 |
ENSDART00000054479
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr19_+_9293058 | 0.15 |
ENSDART00000146218
|
ndufv1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1 |
| chr8_-_49227143 | 0.15 |
|
|
|
| chr2_-_41722058 | 0.14 |
ENSDART00000022643
|
znf622
|
zinc finger protein 622 |
| chr17_+_8018808 | 0.13 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr10_+_20222743 | 0.13 |
ENSDART00000167008
|
ppp3ccb
|
protein phosphatase 3, catalytic subunit, gamma isozyme, b |
| chr25_+_32419062 | 0.12 |
|
|
|
| chr10_-_15090551 | 0.10 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr7_-_20211716 | 0.09 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr17_+_24585915 | 0.08 |
ENSDART00000082193
|
zmpste24
|
zinc metallopeptidase, STE24 homolog |
| chr10_-_35164761 | 0.08 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr15_-_24007354 | 0.08 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr15_+_31956131 | 0.07 |
|
|
|
| chr3_-_13311188 | 0.07 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr4_-_16002407 | 0.07 |
ENSDART00000125389
|
mest
|
mesoderm specific transcript |
| chr20_+_53772078 | 0.06 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr8_+_7815304 | 0.06 |
ENSDART00000165575
|
cxxc1a
|
CXXC finger protein 1a |
| chr24_-_26334771 | 0.05 |
ENSDART00000079984
ENSDART00000136871 |
rpl22l1
|
ribosomal protein L22-like 1 |
| chr21_-_27158568 | 0.04 |
ENSDART00000065401
|
zgc:110782
|
zgc:110782 |
| chr20_+_53772470 | 0.04 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr12_-_26323696 | 0.02 |
|
|
|
| chr10_+_26691086 | 0.02 |
ENSDART00000079174
|
htatsf1
|
HIV-1 Tat specific factor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.3 | 1.8 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 0.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.5 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 | 0.8 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.2 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 2.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 1.1 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.1 | GO:0055004 | atrial cardiac myofibril assembly(GO:0055004) |
| 0.0 | 0.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.5 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.6 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.0 | GO:0005813 | centrosome(GO:0005813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 0.5 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.5 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.4 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.1 | 0.2 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 1.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.6 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 3.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.6 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 0.8 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |