Project
DANIO-CODE
Navigation
Downloads
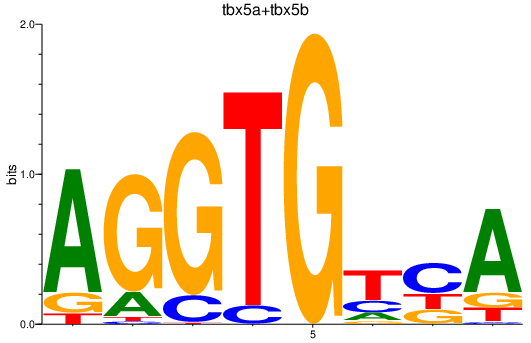
Results for tbx5a+tbx5b
Z-value: 1.17
Transcription factors associated with tbx5a+tbx5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx5a
|
ENSDARG00000024894 | T-box transcription factor 5a |
|
tbx5b
|
ENSDARG00000092060 | T-box transcription factor 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx5a | dr10_dc_chr5_-_71505131_71505211 | -0.62 | 1.0e-02 | Click! |
Activity profile of tbx5a+tbx5b motif
Sorted Z-values of tbx5a+tbx5b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx5a+tbx5b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_35411993 | 4.31 |
ENSDART00000051751
|
zgc:113424
|
zgc:113424 |
| chr19_-_35408237 | 3.44 |
ENSDART00000141704
|
si:rp71-45k5.2
|
si:rp71-45k5.2 |
| chr11_+_29299382 | 2.65 |
|
|
|
| chr24_-_9880951 | 2.56 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr11_-_6442588 | 2.35 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr20_-_14218080 | 2.19 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr7_-_55346967 | 2.16 |
ENSDART00000135304
|
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr24_+_9887575 | 2.13 |
ENSDART00000172773
|
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr24_-_9857510 | 2.12 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr5_-_15971338 | 2.03 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr18_-_14306241 | 1.92 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr24_-_9844547 | 1.89 |
|
|
|
| chr24_-_9865837 | 1.85 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr2_-_38305602 | 1.82 |
ENSDART00000061677
|
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr11_+_29319568 | 1.77 |
ENSDART00000158818
|
BX548000.2
|
ENSDARG00000099044 |
| chr4_+_16020464 | 1.76 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr22_+_25113293 | 1.75 |
ENSDART00000171851
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr13_-_28480530 | 1.72 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr5_-_31738565 | 1.69 |
ENSDART00000017956
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr4_+_14983045 | 1.67 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr23_-_27579257 | 1.66 |
ENSDART00000137229
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr11_+_29290515 | 1.63 |
ENSDART00000103388
|
wu:fi42e03
|
wu:fi42e03 |
| chr11_-_15886860 | 1.59 |
ENSDART00000170731
|
zgc:173544
|
zgc:173544 |
| chr11_-_25615491 | 1.57 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr16_+_10373338 | 1.45 |
ENSDART00000091377
|
mrs2
|
MRS2 magnesium transporter |
| chr6_-_25065823 | 1.44 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr22_-_17627900 | 1.44 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr5_+_61711614 | 1.41 |
ENSDART00000082965
|
ABR
|
active BCR-related |
| chr10_-_45182378 | 1.36 |
ENSDART00000161815
|
polm
|
polymerase (DNA directed), mu |
| chr23_-_29886117 | 1.34 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr21_-_30217360 | 1.33 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr5_-_31796376 | 1.32 |
ENSDART00000142095
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr21_-_3548863 | 1.30 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr19_-_35411924 | 1.29 |
ENSDART00000051751
|
zgc:113424
|
zgc:113424 |
| chr17_-_31387062 | 1.26 |
|
|
|
| chr14_-_33641145 | 1.26 |
ENSDART00000167774
|
foxa
|
forkhead box A sequence |
| chr5_+_29052293 | 1.24 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr23_+_19663746 | 1.24 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr23_-_10810190 | 1.23 |
ENSDART00000140745
|
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr18_+_18077901 | 1.22 |
|
|
|
| chr5_+_56988663 | 1.21 |
ENSDART00000074268
|
zgc:153929
|
zgc:153929 |
| chr10_+_37457234 | 1.20 |
ENSDART00000136554
|
akap1a
|
A kinase (PRKA) anchor protein 1a |
| chr1_+_21964075 | 1.20 |
ENSDART00000161874
|
anxa5b
|
annexin A5b |
| chr8_-_38284959 | 1.19 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr15_-_25333773 | 1.19 |
ENSDART00000078230
|
mettl16
|
methyltransferase like 16 |
| chr24_+_15510787 | 1.18 |
ENSDART00000042943
|
fbxo15
|
F-box protein 15 |
| chr23_-_34017934 | 1.18 |
ENSDART00000133223
|
ENSDARG00000070393
|
ENSDARG00000070393 |
| chr15_+_40232397 | 1.17 |
ENSDART00000154947
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr19_-_8893920 | 1.17 |
ENSDART00000025385
|
cers2a
|
ceramide synthase 2a |
| chr9_-_28444272 | 1.17 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr2_+_36111019 | 1.15 |
ENSDART00000161694
|
BX681417.4
|
ENSDARG00000101668 |
| chr14_+_50263527 | 1.15 |
ENSDART00000126260
|
anxa6
|
annexin A6 |
| chr15_-_43283562 | 1.13 |
ENSDART00000110352
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr3_+_40113121 | 1.13 |
ENSDART00000074746
|
smcr8a
|
Smith-Magenis syndrome chromosome region, candidate 8a |
| chr15_-_25333252 | 1.12 |
ENSDART00000127771
|
mettl16
|
methyltransferase like 16 |
| chr13_-_28480153 | 1.11 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr3_+_59972297 | 1.11 |
|
|
|
| chr5_+_58936399 | 1.10 |
|
|
|
| chr3_+_51308904 | 1.09 |
ENSDART00000008607
|
ttyh2l
|
tweety homolog 2, like |
| chr13_-_7972530 | 1.07 |
ENSDART00000080460
|
atl2
|
atlastin GTPase 2 |
| chr9_-_4640050 | 1.06 |
ENSDART00000171634
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr6_-_25065745 | 1.05 |
ENSDART00000165170
|
znf326
|
zinc finger protein 326 |
| chr5_-_31796734 | 1.05 |
ENSDART00000142095
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr3_-_54352535 | 1.05 |
ENSDART00000021977
ENSDART00000078973 |
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr1_+_49170967 | 1.04 |
ENSDART00000164936
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr13_-_7972442 | 1.04 |
ENSDART00000128005
|
atl2
|
atlastin GTPase 2 |
| chr20_+_9136578 | 1.04 |
ENSDART00000064150
ENSDART00000158196 |
ENSDARG00000043687
|
ENSDARG00000043687 |
| chr11_-_40239946 | 1.03 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr8_-_38284748 | 1.02 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr22_-_17627831 | 1.00 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr17_+_15666078 | 0.98 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr3_-_31942015 | 0.97 |
|
|
|
| chr2_+_9762781 | 0.96 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr15_-_16241412 | 0.96 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr10_-_24350114 | 0.96 |
ENSDART00000109549
ENSDART00000148480 |
inpp5kb
|
inositol polyphosphate-5-phosphatase Kb |
| chr23_+_37476457 | 0.96 |
ENSDART00000178064
|
AL954146.3
|
ENSDARG00000108629 |
| chr22_+_26840517 | 0.95 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr4_+_4825461 | 0.94 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr9_-_9864149 | 0.94 |
ENSDART00000121456
|
fstl1b
|
follistatin-like 1b |
| chr8_+_13327234 | 0.92 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr2_+_10040130 | 0.91 |
ENSDART00000139639
|
anxa13l
|
annexin A13, like |
| chr19_+_791656 | 0.90 |
ENSDART00000138406
|
tmem79a
|
transmembrane protein 79a |
| chr2_-_23735018 | 0.89 |
ENSDART00000020136
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr7_+_36196295 | 0.89 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr17_-_24860924 | 0.89 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr5_+_25474996 | 0.87 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr6_-_8225364 | 0.87 |
|
|
|
| chr9_-_28444024 | 0.87 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr14_-_40951773 | 0.87 |
|
|
|
| chr15_-_1519615 | 0.86 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr7_-_20556760 | 0.85 |
ENSDART00000143509
|
dnajc3b
|
DnaJ (Hsp40) homolog, subfamily C, member 3b |
| chr14_+_7391258 | 0.85 |
ENSDART00000162363
|
brd8
|
bromodomain containing 8 |
| chr7_+_13238684 | 0.85 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr2_-_57502821 | 0.85 |
|
|
|
| chr19_+_34263554 | 0.85 |
ENSDART00000161290
|
pex1
|
peroxisomal biogenesis factor 1 |
| chr12_+_29121368 | 0.85 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr9_-_29128379 | 0.85 |
ENSDART00000101269
|
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr14_+_27984076 | 0.84 |
ENSDART00000173237
|
mid2
|
midline 2 |
| chr8_-_13231600 | 0.84 |
ENSDART00000014528
|
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr7_+_48025010 | 0.84 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr7_+_13742576 | 0.83 |
|
|
|
| chr21_-_30217568 | 0.83 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr1_+_22160932 | 0.83 |
ENSDART00000016488
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr2_-_45657620 | 0.82 |
ENSDART00000056357
|
gpsm2
|
G protein signaling modulator 2 |
| chr23_-_41367783 | 0.82 |
|
|
|
| chr3_+_58417512 | 0.82 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr12_+_33359784 | 0.82 |
ENSDART00000007053
|
narf
|
nuclear prelamin A recognition factor |
| chr20_-_43846604 | 0.82 |
ENSDART00000150078
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr5_+_12971972 | 0.81 |
ENSDART00000132406
|
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr21_-_14154489 | 0.81 |
ENSDART00000114715
|
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr11_+_12754166 | 0.80 |
ENSDART00000123445
ENSDART00000163364 ENSDART00000066122 |
rtel1
|
regulator of telomere elongation helicase 1 |
| chr8_+_3373066 | 0.80 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr8_+_20108592 | 0.80 |
|
|
|
| chr2_-_56632969 | 0.79 |
ENSDART00000089158
|
hmha1a
|
histocompatibility (minor) HA-1 a |
| chr21_+_20350218 | 0.79 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr24_-_32259029 | 0.79 |
ENSDART00000048633
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr23_-_10810264 | 0.78 |
ENSDART00000013768
|
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr24_+_15995931 | 0.78 |
ENSDART00000105955
|
TIMM21
|
translocase of inner mitochondrial membrane 21 |
| chr7_+_38690837 | 0.78 |
ENSDART00000173565
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr3_+_44928592 | 0.77 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr8_-_38284904 | 0.77 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr1_+_40428827 | 0.77 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr9_+_27909662 | 0.76 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr1_+_48770997 | 0.76 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr11_+_24756894 | 0.75 |
ENSDART00000129211
|
ENSDARG00000044874
|
ENSDARG00000044874 |
| chr13_-_28480365 | 0.74 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr24_+_15510895 | 0.74 |
ENSDART00000143160
|
fbxo15
|
F-box protein 15 |
| chr1_+_49171052 | 0.74 |
ENSDART00000164936
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr10_-_24350023 | 0.73 |
ENSDART00000109549
ENSDART00000148480 |
inpp5kb
|
inositol polyphosphate-5-phosphatase Kb |
| chr3_-_40090673 | 0.73 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr5_+_60234640 | 0.73 |
|
|
|
| chr15_+_21776031 | 0.72 |
ENSDART00000136151
|
NKAPD1
|
zgc:162339 |
| chr19_-_6164713 | 0.72 |
ENSDART00000082012
|
gsk3aa
|
glycogen synthase kinase 3 alpha a |
| chr23_+_32015527 | 0.71 |
ENSDART00000088607
|
nemp1
|
nuclear envelope integral membrane protein 1 |
| chr12_+_6197452 | 0.71 |
|
|
|
| chr3_-_40112966 | 0.71 |
ENSDART00000154562
|
top3a
|
topoisomerase (DNA) III alpha |
| chr11_+_24562450 | 0.70 |
ENSDART00000082761
ENSDART00000131976 |
adipor1a
|
adiponectin receptor 1a |
| chr14_+_23631214 | 0.70 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr1_+_14572155 | 0.69 |
ENSDART00000033018
|
pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr10_+_42841562 | 0.69 |
|
|
|
| chr5_+_25408121 | 0.69 |
|
|
|
| chr21_-_36710989 | 0.69 |
ENSDART00000086060
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr12_+_18407380 | 0.69 |
ENSDART00000146260
|
msrb1b
|
methionine sulfoxide reductase B1b |
| chr2_-_57502700 | 0.68 |
|
|
|
| chr12_+_18407435 | 0.68 |
ENSDART00000146260
|
msrb1b
|
methionine sulfoxide reductase B1b |
| chr4_-_16611960 | 0.67 |
|
|
|
| chr5_-_35997345 | 0.66 |
ENSDART00000122098
|
rhogc
|
ras homolog gene family, member Gc |
| chr21_-_35291005 | 0.66 |
ENSDART00000134780
|
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr1_+_49170632 | 0.66 |
ENSDART00000132405
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr13_-_24778226 | 0.66 |
|
|
|
| chr2_-_32278726 | 0.66 |
ENSDART00000159843
ENSDART00000056621 ENSDART00000039717 ENSDART00000145704 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr15_+_40228295 | 0.66 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr20_+_36957917 | 0.65 |
ENSDART00000153091
|
BX248390.1
|
ENSDARG00000096735 |
| chr4_+_9477541 | 0.65 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr8_+_3372903 | 0.65 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr21_-_19883182 | 0.65 |
ENSDART00000065670
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr5_+_61711709 | 0.65 |
ENSDART00000082965
|
ABR
|
active BCR-related |
| chr7_-_48394268 | 0.65 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr2_+_9762627 | 0.64 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr10_-_20629538 | 0.64 |
|
|
|
| chr1_-_55211232 | 0.64 |
ENSDART00000064194
|
asf1bb
|
anti-silencing function 1Bb histone chaperone |
| chr7_+_26274744 | 0.64 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr20_+_45585743 | 0.63 |
|
|
|
| chr5_+_29052517 | 0.63 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr15_-_16241500 | 0.62 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr15_-_25333737 | 0.62 |
ENSDART00000078230
|
mettl16
|
methyltransferase like 16 |
| chr10_-_31129270 | 0.62 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr16_+_4183457 | 0.62 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr10_+_6339059 | 0.62 |
ENSDART00000160813
|
tpm2
|
tropomyosin 2 (beta) |
| chr23_-_29886214 | 0.61 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr3_-_39163225 | 0.61 |
ENSDART00000102674
ENSDART00000167070 |
plcd3a
|
phospholipase C, delta 3a |
| chr21_-_34938261 | 0.61 |
ENSDART00000023838
|
lipia
|
lipase, member Ia |
| chr21_+_20350334 | 0.61 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr10_-_24350068 | 0.60 |
ENSDART00000109549
ENSDART00000148480 |
inpp5kb
|
inositol polyphosphate-5-phosphatase Kb |
| chr4_-_16611908 | 0.60 |
|
|
|
| chr23_+_4315077 | 0.59 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr6_+_8417443 | 0.59 |
|
|
|
| chr10_-_35205636 | 0.59 |
ENSDART00000131291
|
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr20_-_54285455 | 0.58 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr13_-_24778352 | 0.57 |
|
|
|
| chr16_-_12206435 | 0.57 |
ENSDART00000103944
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr6_+_27523853 | 0.57 |
ENSDART00000128985
|
ryk
|
receptor-like tyrosine kinase |
| chr7_+_26274687 | 0.57 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr5_-_26293054 | 0.57 |
ENSDART00000126669
|
lman2lb
|
lectin, mannose-binding 2-like b |
| chr14_-_46599151 | 0.56 |
|
|
|
| chr23_-_31339986 | 0.56 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr23_+_16822435 | 0.55 |
ENSDART00000137737
|
fbxo44
|
F-box protein 44 |
| chr18_-_20832736 | 0.55 |
ENSDART00000100743
|
cttnbp2
|
cortactin binding protein 2 |
| chr1_+_40428722 | 0.55 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr19_+_37548595 | 0.55 |
ENSDART00000103159
|
smim12
|
small integral membrane protein 12 |
| chr20_+_35176069 | 0.54 |
|
|
|
| chr9_-_4640088 | 0.54 |
ENSDART00000179110
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr8_+_29733037 | 0.54 |
ENSDART00000133955
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr22_+_18294579 | 0.54 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr11_-_25615683 | 0.53 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr1_-_529071 | 0.53 |
ENSDART00000147610
|
si:ch73-41e3.7
|
si:ch73-41e3.7 |
| chr7_+_58541247 | 0.53 |
ENSDART00000165852
|
lypla1
|
lysophospholipase I |
| chr13_-_24778261 | 0.52 |
|
|
|
| chr6_+_9735100 | 0.52 |
ENSDART00000171518
|
abi2b
|
abl-interactor 2b |
| chr5_+_25475041 | 0.52 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:1903449 | male sex determination(GO:0030238) androst-4-ene-3,17-dione biosynthetic process(GO:1903449) |
| 0.7 | 2.0 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.6 | 1.9 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.6 | 1.7 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.5 | 2.3 | GO:0045719 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.4 | 2.4 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.4 | 2.8 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.3 | 1.3 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.3 | 2.4 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.3 | 1.1 | GO:0003418 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.3 | 0.8 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.2 | 2.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.3 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.2 | 1.5 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.2 | 1.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 1.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 0.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 0.8 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.2 | 2.3 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.2 | 0.5 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.2 | 0.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 0.9 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.2 | 0.6 | GO:0042373 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) vitamin K metabolic process(GO:0042373) |
| 0.2 | 0.6 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.8 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.1 | 1.2 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.4 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.4 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.3 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 1.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.4 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 1.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 1.8 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 1.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.5 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.8 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 2.2 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 1.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 3.7 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.6 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.3 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.7 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.7 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.4 | GO:0046651 | mononuclear cell proliferation(GO:0032943) T cell proliferation(GO:0042098) lymphocyte proliferation(GO:0046651) leukocyte proliferation(GO:0070661) |
| 0.1 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 1.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 2.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 0.5 | GO:0061458 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.5 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:1990092 | calcium-dependent self proteolysis(GO:1990092) |
| 0.0 | 0.9 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.2 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.5 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.7 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0055004 | atrial cardiac myofibril assembly(GO:0055004) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.2 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 1.5 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.8 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.2 | GO:0070252 | actin-mediated cell contraction(GO:0070252) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.0 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.3 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.8 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.1 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 0.3 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.7 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 1.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.2 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.8 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.3 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.9 | GO:0045055 | regulated exocytosis(GO:0045055) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.6 | 2.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.4 | 1.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 1.5 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.3 | 2.0 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 0.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 2.0 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 2.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.3 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 2.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 2.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 2.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.2 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.9 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0052907 | U6 snRNA 3'-end binding(GO:0030629) 23S rRNA (adenine(1618)-N(6))-methyltransferase activity(GO:0052907) |
| 0.9 | 3.6 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.6 | 2.3 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.5 | 2.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.5 | 1.4 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.3 | 0.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 0.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.3 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 1.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.2 | 1.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.9 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.2 | 0.6 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.2 | 1.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.5 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 1.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 2.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 3.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 2.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 2.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 2.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.7 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.8 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 1.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.3 | GO:0008465 | glycerate dehydrogenase activity(GO:0008465) |
| 0.1 | 0.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 2.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.6 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 0.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 3.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.7 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 2.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.2 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.1 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.6 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.3 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 2.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.9 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 1.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 4.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 3.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 4.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 1.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 3.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 2.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 2.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |