Project
DANIO-CODE
Navigation
Downloads
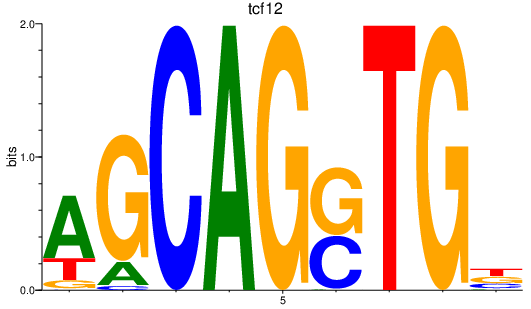
Results for tcf12
Z-value: 1.67
Transcription factors associated with tcf12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf12
|
ENSDARG00000004714 | transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf12 | dr10_dc_chr7_-_52434786_52434823 | 0.86 | 2.0e-05 | Click! |
Activity profile of tcf12 motif
Sorted Z-values of tcf12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_25659535 | 4.77 |
ENSDART00000135627
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr2_-_16896965 | 4.32 |
ENSDART00000022549
|
atp1b3a
|
ATPase, Na+/K+ transporting, beta 3a polypeptide |
| chr19_-_30816780 | 4.18 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr19_+_31046291 | 3.96 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr10_+_15819127 | 3.91 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr4_-_14330113 | 3.84 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr7_+_49442972 | 3.80 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr18_-_11216447 | 3.69 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr7_+_6517248 | 3.50 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr15_-_15421065 | 3.36 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr2_-_30675594 | 3.28 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr11_-_25803101 | 3.27 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr18_+_62936 | 3.20 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr10_+_33227967 | 3.20 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr15_+_34131126 | 3.19 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr8_-_985673 | 3.12 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr24_-_33817169 | 3.08 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr18_-_3244618 | 3.03 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr9_-_23081918 | 3.02 |
ENSDART00000143888
|
neb
|
nebulin |
| chr7_+_40972616 | 3.02 |
ENSDART00000173814
|
scrib
|
scribbled planar cell polarity protein |
| chr7_+_39173765 | 2.92 |
ENSDART00000173748
|
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr6_-_39315024 | 2.92 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr22_-_37468331 | 2.91 |
ENSDART00000160940
|
SOX2OT_exon1
|
SOX2 overlapping transcript exon 1 |
| chr8_-_7049946 | 2.89 |
ENSDART00000045669
ENSDART00000162950 |
KRT73
|
keratin 73 |
| chr10_-_43826919 | 2.88 |
ENSDART00000039551
ENSDART00000160786 ENSDART00000099134 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr25_-_2485276 | 2.83 |
ENSDART00000154889
ENSDART00000155027 |
BX957346.1
|
ENSDARG00000096850 |
| chr2_-_21693984 | 2.82 |
ENSDART00000171699
|
hhatla
|
hedgehog acyltransferase-like, a |
| chr23_+_21546553 | 2.81 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr23_+_24141576 | 2.75 |
|
|
|
| chr5_-_40133824 | 2.71 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr18_+_38307946 | 2.68 |
ENSDART00000134247
|
lmo2
|
LIM domain only 2 (rhombotin-like 1) |
| chr17_+_23278879 | 2.67 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr10_+_31358236 | 2.66 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr10_-_10372266 | 2.66 |
|
|
|
| chr3_-_48865474 | 2.65 |
ENSDART00000133036
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr22_+_26543146 | 2.65 |
|
|
|
| chr24_+_17125429 | 2.62 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr20_-_32543497 | 2.62 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr4_+_12618256 | 2.60 |
ENSDART00000112860
ENSDART00000134362 |
lmo3
|
LIM domain only 3 |
| chr23_-_7892862 | 2.58 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr9_-_23081250 | 2.55 |
|
|
|
| chr9_+_308260 | 2.54 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr21_+_23916512 | 2.53 |
ENSDART00000145541
ENSDART00000065599 ENSDART00000112869 |
cadm1a
|
cell adhesion molecule 1a |
| chr14_+_21531709 | 2.50 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr14_-_36523075 | 2.49 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr8_-_44305494 | 2.49 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr23_-_27226280 | 2.49 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr16_+_5625301 | 2.48 |
|
|
|
| chr19_-_23037220 | 2.47 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr3_+_32394653 | 2.45 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr12_-_5085227 | 2.42 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr14_+_34683602 | 2.40 |
ENSDART00000172171
|
ebf1a
|
early B-cell factor 1a |
| chr9_+_42019770 | 2.40 |
ENSDART00000097295
|
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr18_+_9413588 | 2.39 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr24_+_9605832 | 2.35 |
ENSDART00000131891
|
tmem108
|
transmembrane protein 108 |
| chr5_-_36647498 | 2.34 |
ENSDART00000112312
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr3_+_37433008 | 2.32 |
ENSDART00000055225
|
wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr9_-_7560915 | 2.32 |
ENSDART00000081553
|
desma
|
desmin a |
| chr11_-_41357639 | 2.31 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr7_+_6814828 | 2.30 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr22_-_17652938 | 2.28 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr7_+_21455138 | 2.28 |
ENSDART00000173992
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr3_-_55956030 | 2.26 |
ENSDART00000158163
|
srl
|
sarcalumenin |
| chr4_+_17428131 | 2.24 |
ENSDART00000056005
|
ascl1a
|
achaete-scute family bHLH transcription factor 1a |
| KN149968v1_+_15749 | 2.24 |
ENSDART00000168977
|
ENSDARG00000102503
|
ENSDARG00000102503 |
| chr9_-_34491458 | 2.20 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| KN149932v1_+_27584 | 2.19 |
|
|
|
| chr2_-_20941256 | 2.17 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr5_+_45477559 | 2.17 |
ENSDART00000138055
|
iqgap2
|
IQ motif containing GTPase activating protein 2 |
| chr1_-_54319943 | 2.17 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr12_-_36138709 | 2.16 |
ENSDART00000130985
|
st6galnac
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase |
| chr10_-_39210658 | 2.16 |
ENSDART00000150193
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr4_+_10065500 | 2.16 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr10_-_42032702 | 2.16 |
|
|
|
| chr20_-_19646761 | 2.15 |
|
|
|
| chr24_-_3334678 | 2.13 |
|
|
|
| chr13_-_31310439 | 2.13 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr23_-_39956151 | 2.13 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr21_+_27346176 | 2.09 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr1_-_10822278 | 2.08 |
ENSDART00000091205
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr16_+_43249142 | 2.07 |
ENSDART00000154493
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr10_+_15819085 | 2.07 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr5_-_28090077 | 2.06 |
|
|
|
| chr11_+_14142126 | 2.05 |
ENSDART00000102520
|
palm1a
|
paralemmin 1a |
| chr25_+_35179025 | 2.05 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr14_+_11151485 | 2.04 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr5_-_36237656 | 2.03 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr11_-_5868257 | 2.02 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr5_+_36180981 | 2.02 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr11_+_20269868 | 2.01 |
ENSDART00000104022
|
cdh4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr16_+_24057260 | 2.00 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr16_+_46435014 | 1.99 |
ENSDART00000144000
|
rpz2
|
rapunzel 2 |
| chr21_-_17259886 | 1.99 |
ENSDART00000114877
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr11_+_666006 | 1.98 |
ENSDART00000130839
|
timp4.1
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 1 |
| chr17_-_6219019 | 1.98 |
|
|
|
| chr23_-_31585883 | 1.98 |
ENSDART00000157511
|
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr9_+_9133904 | 1.98 |
ENSDART00000165932
|
sik1
|
salt-inducible kinase 1 |
| chr16_-_20506304 | 1.97 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr14_+_26247858 | 1.97 |
ENSDART00000105932
|
si:dkeyp-110e4.11
|
si:dkeyp-110e4.11 |
| chr22_+_4722336 | 1.97 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr8_-_23172938 | 1.93 |
ENSDART00000141090
|
si:ch211-196c10.15
|
si:ch211-196c10.15 |
| chr22_-_613948 | 1.90 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr16_+_5625552 | 1.90 |
|
|
|
| chr15_-_35563367 | 1.88 |
|
|
|
| chr15_+_9351511 | 1.88 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr16_+_3090170 | 1.88 |
ENSDART00000110395
|
limd1a
|
LIM domains containing 1a |
| chr22_+_12327849 | 1.88 |
ENSDART00000178678
|
FO704572.1
|
ENSDARG00000106890 |
| chr16_-_42040346 | 1.88 |
ENSDART00000169313
|
pycard
|
PYD and CARD domain containing |
| chr21_+_4964891 | 1.86 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr19_-_22182031 | 1.86 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr2_-_44402486 | 1.86 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr24_-_2278409 | 1.85 |
|
|
|
| chr3_-_38550961 | 1.84 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr16_-_26803204 | 1.83 |
ENSDART00000162665
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr23_-_15807415 | 1.82 |
ENSDART00000178557
|
BX005011.3
|
ENSDARG00000108083 |
| chr3_-_39313029 | 1.81 |
ENSDART00000156123
|
CU464118.1
|
ENSDARG00000097747 |
| chr15_-_1858350 | 1.81 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr16_-_14463682 | 1.81 |
ENSDART00000011224
|
itga10
|
integrin, alpha 10 |
| chr23_+_20183765 | 1.80 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr20_-_42805703 | 1.80 |
ENSDART00000045816
|
plg
|
plasminogen |
| chr6_-_16541104 | 1.79 |
|
|
|
| chr1_-_48853800 | 1.79 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr21_-_34623741 | 1.79 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr11_-_9479592 | 1.79 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr18_-_15404998 | 1.78 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr2_-_42643045 | 1.78 |
ENSDART00000139929
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr13_-_31165867 | 1.78 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr17_+_8166167 | 1.77 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr12_-_4767887 | 1.76 |
ENSDART00000167490
|
mapta
|
microtubule-associated protein tau a |
| chr20_-_22036656 | 1.75 |
ENSDART00000152290
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr25_+_14411153 | 1.75 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr14_-_33596430 | 1.74 |
ENSDART00000112438
|
si:ch73-335m24.5
|
si:ch73-335m24.5 |
| chr13_-_24181106 | 1.74 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr23_+_36032206 | 1.73 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr23_-_27226387 | 1.73 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr24_-_33817036 | 1.73 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr4_+_12618485 | 1.73 |
ENSDART00000112860
|
lmo3
|
LIM domain only 3 |
| chr5_+_53854279 | 1.72 |
ENSDART00000165889
|
tprn
|
taperin |
| chr7_+_31608828 | 1.72 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr6_-_586251 | 1.71 |
ENSDART00000148867
ENSDART00000149414 ENSDART00000149248 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr20_+_34497817 | 1.71 |
ENSDART00000061632
|
fam129aa
|
family with sequence similarity 129, member Aa |
| chr10_-_36462538 | 1.71 |
ENSDART00000161823
|
ubl3a
|
ubiquitin-like 3a |
| chr10_+_16634200 | 1.71 |
ENSDART00000101142
|
chsy3
|
chondroitin sulfate synthase 3 |
| chr4_-_4825948 | 1.70 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr18_-_34166500 | 1.70 |
ENSDART00000079341
|
plch1
|
phospholipase C, eta 1 |
| chr10_-_10374032 | 1.70 |
ENSDART00000134929
|
BX323065.1
|
ENSDARG00000095365 |
| chr4_-_14330048 | 1.69 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr6_+_1873957 | 1.69 |
|
|
|
| chr11_-_6960107 | 1.69 |
ENSDART00000171255
|
COMP
|
cartilage oligomeric matrix protein |
| chr5_-_23697487 | 1.69 |
ENSDART00000013309
|
sox19a
|
SRY (sex determining region Y)-box 19a |
| KN150178v1_-_23644 | 1.68 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr16_+_23172295 | 1.67 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr14_-_2027432 | 1.67 |
|
|
|
| chr1_+_38834751 | 1.66 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr12_-_25825072 | 1.66 |
|
|
|
| chr12_+_22734974 | 1.66 |
ENSDART00000130594
|
afap1
|
actin filament associated protein 1 |
| chr24_+_20414578 | 1.66 |
ENSDART00000131677
|
hhatlb
|
hedgehog acyltransferase-like, b |
| chr1_+_51925783 | 1.65 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr6_-_32718634 | 1.65 |
ENSDART00000175666
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr14_-_16023198 | 1.64 |
|
|
|
| chr9_-_49834232 | 1.64 |
|
|
|
| chr23_+_14058972 | 1.63 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr15_+_28435937 | 1.63 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr22_+_21997000 | 1.62 |
ENSDART00000046174
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr11_-_30105047 | 1.62 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr14_+_36156947 | 1.61 |
|
|
|
| KN150495v1_-_20497 | 1.60 |
|
|
|
| chr9_-_32942783 | 1.60 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr23_+_6861489 | 1.60 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr23_+_36002332 | 1.60 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr16_+_28819826 | 1.60 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr22_+_38096040 | 1.60 |
ENSDART00000097533
|
wwtr1
|
WW domain containing transcription regulator 1 |
| chr14_+_38445969 | 1.60 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr9_-_18561215 | 1.59 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr21_-_5852663 | 1.58 |
ENSDART00000130521
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr3_-_27944852 | 1.58 |
ENSDART00000122037
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr22_+_24131239 | 1.58 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr2_-_30071872 | 1.58 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr7_+_16583234 | 1.57 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr8_+_23464087 | 1.57 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr25_+_24193604 | 1.56 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr23_-_3466041 | 1.56 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr6_+_59762717 | 1.55 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr15_+_7190427 | 1.55 |
ENSDART00000109394
|
her13
|
hairy-related 13 |
| chr5_+_38237115 | 1.55 |
ENSDART00000160236
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr16_+_24032160 | 1.55 |
ENSDART00000103190
|
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr4_+_8637319 | 1.55 |
|
|
|
| chr10_-_39211281 | 1.55 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr22_+_16282153 | 1.54 |
ENSDART00000162685
ENSDART00000105678 |
lrrc39
|
leucine rich repeat containing 39 |
| chr1_-_16894589 | 1.53 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long-chain family member 1a |
| chr21_-_7277655 | 1.53 |
ENSDART00000056561
|
s100z
|
S100 calcium binding protein Z |
| chr2_+_21342233 | 1.53 |
ENSDART00000062563
|
rreb1b
|
ras responsive element binding protein 1b |
| chr4_+_21166170 | 1.53 |
ENSDART00000036886
|
nav3
|
neuron navigator 3 |
| chr25_-_13737344 | 1.52 |
|
|
|
| chr7_+_31567166 | 1.52 |
ENSDART00000099785
ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr11_-_25592102 | 1.52 |
ENSDART00000103549
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr6_+_45917081 | 1.52 |
ENSDART00000149450
ENSDART00000149642 |
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr10_+_34377959 | 1.52 |
|
|
|
| chr14_+_34683695 | 1.51 |
ENSDART00000170631
|
ebf1a
|
early B-cell factor 1a |
| chr5_+_37053530 | 1.51 |
ENSDART00000161051
|
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0060262 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 1.1 | 4.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 1.1 | 6.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 1.1 | 3.2 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 1.0 | 2.9 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.8 | 2.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.8 | 2.4 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.8 | 3.8 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.8 | 3.0 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.7 | 2.2 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.7 | 8.1 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.7 | 2.8 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.7 | 7.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.7 | 2.1 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.7 | 2.0 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.7 | 2.0 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.6 | 2.6 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.6 | 1.9 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.6 | 1.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.6 | 1.7 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.6 | 2.2 | GO:0032371 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.5 | 2.1 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.5 | 1.6 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.5 | 1.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.5 | 3.0 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.5 | 3.0 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.5 | 2.8 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.5 | 1.9 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 1.3 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.4 | 1.8 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.4 | 0.4 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.4 | 1.3 | GO:0030431 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.4 | 1.3 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.4 | 1.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.4 | 2.3 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.4 | 1.5 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.4 | 1.1 | GO:0097186 | enamel mineralization(GO:0070166) amelogenesis(GO:0097186) |
| 0.4 | 4.8 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.4 | 0.7 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.3 | 2.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.3 | 3.7 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.3 | 4.3 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.3 | 1.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.3 | 1.3 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.3 | 2.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.3 | 2.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 1.9 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.3 | 1.6 | GO:0090200 | regulation of mitochondrial membrane potential(GO:0051881) membrane depolarization(GO:0051899) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.3 | 1.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 1.6 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.3 | 1.8 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.3 | 2.4 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.3 | 0.9 | GO:0021661 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.3 | 0.9 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 1.8 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.3 | 1.5 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 0.9 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.3 | 1.1 | GO:0071635 | transforming growth factor beta activation(GO:0036363) transforming growth factor beta production(GO:0071604) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.3 | 1.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 1.7 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.3 | 3.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.3 | 1.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.3 | 1.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 2.4 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.3 | 1.9 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.3 | 1.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 0.8 | GO:0030800 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.3 | 3.7 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 0.8 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.3 | 2.0 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.3 | 0.8 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 0.7 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.2 | 1.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 1.5 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 1.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.2 | 1.9 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.2 | 0.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 1.8 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 1.3 | GO:0034672 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.2 | 4.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 1.2 | GO:0001964 | startle response(GO:0001964) |
| 0.2 | 4.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 0.6 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 2.9 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.2 | 1.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.2 | 0.2 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.2 | 1.3 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 2.6 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 0.6 | GO:0050810 | positive regulation of steroid metabolic process(GO:0045940) regulation of steroid biosynthetic process(GO:0050810) |
| 0.2 | 1.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 0.9 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.2 | 1.9 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.2 | 1.0 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 3.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 0.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.2 | 1.7 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.2 | 1.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 0.7 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.2 | 4.9 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.2 | 0.5 | GO:0097378 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.2 | 2.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 0.5 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.2 | 3.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 2.1 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.2 | 0.9 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.2 | 1.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.2 | 0.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.2 | 2.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.1 | GO:0019374 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) short-chain fatty acid metabolic process(GO:0046459) |
| 0.2 | 0.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.9 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 0.8 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.2 | 3.3 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 1.5 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 0.6 | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 0.6 | GO:0072506 | regulation of fat cell differentiation(GO:0045598) phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.2 | 0.8 | GO:1901381 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.2 | 1.2 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 1.0 | GO:1902866 | regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 1.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.4 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 0.4 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 3.1 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 7.9 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.1 | 2.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.3 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 2.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.9 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 12.5 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 1.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 1.5 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 1.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.4 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.6 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.6 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.4 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.5 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 0.5 | GO:0035908 | ventral aorta development(GO:0035908) pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.1 | 1.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 2.2 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 1.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.5 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.6 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 1.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.5 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 4.7 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 1.9 | GO:0097202 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) activation of cysteine-type endopeptidase activity(GO:0097202) |
| 0.1 | 0.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 2.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.5 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.8 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.6 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 0.4 | GO:0030517 | negative regulation of axon extension(GO:0030517) negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 1.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 5.9 | GO:0021782 | glial cell development(GO:0021782) |
| 0.1 | 1.7 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 1.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.2 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.1 | 0.3 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.1 | 4.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 1.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.9 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 2.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 1.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.4 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.6 | GO:0070252 | actin-mediated cell contraction(GO:0070252) |
| 0.1 | 0.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.2 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.1 | 0.5 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.2 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.1 | 2.1 | GO:0051693 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.1 | 0.6 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.1 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.7 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.1 | 0.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.6 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 1.0 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 2.5 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 1.7 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 1.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0006582 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.7 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 6.0 | GO:0007268 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 1.4 | GO:1990266 | granulocyte migration(GO:0097530) neutrophil migration(GO:1990266) |
| 0.0 | 1.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.4 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.2 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.7 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.6 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 1.1 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 2.2 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 1.2 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 2.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.5 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 1.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.3 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.5 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 1.4 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 1.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.9 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.0 | 1.3 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 1.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.6 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 1.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 1.2 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 1.2 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0071236 | response to antibiotic(GO:0046677) cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 1.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:0051048 | negative regulation of secretion(GO:0051048) negative regulation of secretion by cell(GO:1903531) |
| 0.0 | 1.1 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) positive regulation of oocyte development(GO:0060282) |
| 0.0 | 1.3 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.7 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0048898 | anterior lateral line system development(GO:0048898) anterior lateral line development(GO:0048899) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.3 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.7 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 1.0 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.9 | GO:0051216 | cartilage development(GO:0051216) connective tissue development(GO:0061448) |
| 0.0 | 0.1 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 0.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.4 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 1.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.0 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 0.3 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.1 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.9 | GO:0006869 | lipid transport(GO:0006869) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 1.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.8 | 6.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.6 | 6.2 | GO:0031430 | M band(GO:0031430) |
| 0.6 | 1.7 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.5 | 1.9 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.5 | 2.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 2.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 3.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.3 | 1.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 2.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 0.9 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.3 | 6.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 1.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 2.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 0.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 8.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.2 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 1.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 7.0 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.2 | 2.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 2.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 8.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 4.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 4.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 6.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 6.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.7 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 1.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 3.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 3.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 2.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.0 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 3.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 3.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.7 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 36.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 4.9 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 1.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.5 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 3.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 2.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 5.7 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 2.1 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.1 | 1.4 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 12.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 6.6 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0036477 | somatodendritic compartment(GO:0036477) neuronal cell body(GO:0043025) |
| 0.0 | 1.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 7.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 6.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0070161 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.6 | 1.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.6 | 1.9 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.6 | 2.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 1.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.6 | 4.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 1.6 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.5 | 1.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.5 | 2.4 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.5 | 2.8 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.5 | 6.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 1.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 1.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.4 | 6.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.4 | 5.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.4 | 6.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 3.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.4 | 2.9 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 1.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 1.7 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.3 | 1.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 6.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 4.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 1.5 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.3 | 2.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 0.9 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.3 | 2.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 2.2 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.3 | 1.7 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 1.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 2.0 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 3.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 2.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 4.3 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 5.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.2 | 0.9 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.2 | 2.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 12.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 3.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.2 | 1.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 0.4 | GO:0008556 | potassium-transporting ATPase activity(GO:0008556) |
| 0.2 | 0.6 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 1.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 1.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 0.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 2.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 0.8 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 2.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.9 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 1.2 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.2 | 1.0 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 0.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 1.5 | GO:0051429 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.8 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 0.8 | GO:0017081 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 12.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 2.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.6 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.1 | 1.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.1 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 3.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.6 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 1.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 3.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 1.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 2.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.3 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.1 | 0.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 2.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 3.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 8.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 3.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 23.2 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.2 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 2.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 1.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 2.1 | GO:0005347 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleoside transmembrane transporter activity(GO:0015211) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 3.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 4.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.6 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 2.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 1.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0008235 | metalloexopeptidase activity(GO:0008235) metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.2 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 2.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 48.0 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 2.2 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 1.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 1.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 2.4 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 1.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 4.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 4.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.0 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 1.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 5.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 7.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 2.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 1.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.2 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 11.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.7 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 6.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 0.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 1.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.9 | NABA MATRISOME | Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 3.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 2.4 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.3 | 2.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 2.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 7.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 3.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 1.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 1.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.3 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.1 | 4.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 5.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 4.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.6 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |