Project
DANIO-CODE
Navigation
Downloads
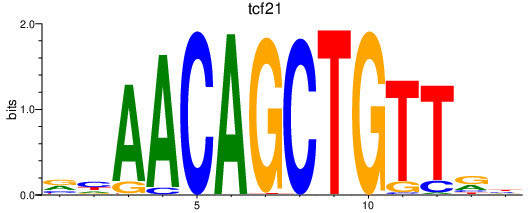
Results for tcf21
Z-value: 0.49
Transcription factors associated with tcf21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf21
|
ENSDARG00000036869 | transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf21 | dr10_dc_chr23_-_31629162_31629165 | -0.74 | 9.3e-04 | Click! |
Activity profile of tcf21 motif
Sorted Z-values of tcf21 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf21
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_20944579 | 0.91 |
ENSDART00000153739
|
nlk1
|
nemo-like kinase, type 1 |
| chr23_-_18131502 | 0.90 |
ENSDART00000173075
|
zgc:92287
|
zgc:92287 |
| chr18_+_14308032 | 0.73 |
|
|
|
| chr8_+_45326435 | 0.71 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr1_-_44892600 | 0.68 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr8_+_25126238 | 0.67 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr11_+_34909167 | 0.63 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr18_-_26815192 | 0.60 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr1_-_33717934 | 0.59 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr21_-_9633519 | 0.59 |
|
|
|
| chr11_+_34909244 | 0.58 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr1_+_46640008 | 0.54 |
ENSDART00000053284
|
bcl9
|
B-cell CLL/lymphoma 9 |
| chr5_+_56809762 | 0.54 |
ENSDART00000135520
|
si:ch211-202f5.3
|
si:ch211-202f5.3 |
| chr15_+_545678 | 0.54 |
ENSDART00000102275
ENSDART00000102274 |
ftr86
|
finTRIM family, member 86 |
| chr2_+_52225622 | 0.54 |
ENSDART00000170353
|
ENSDARG00000103108
|
ENSDARG00000103108 |
| chr23_+_39231080 | 0.53 |
|
|
|
| chr10_-_44713495 | 0.53 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr2_-_41888389 | 0.52 |
|
|
|
| chr10_+_26973377 | 0.51 |
ENSDART00000129993
|
CR450793.1
|
ENSDARG00000088927 |
| chr7_+_34523283 | 0.51 |
ENSDART00000009698
|
esrp2
|
epithelial splicing regulatory protein 2 |
| chr4_+_4825409 | 0.49 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr21_+_22598805 | 0.49 |
ENSDART00000142495
|
BX510940.1
|
ENSDARG00000089088 |
| chr7_+_1337856 | 0.49 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr6_-_8125341 | 0.48 |
ENSDART00000139161
ENSDART00000140021 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr10_+_21477579 | 0.48 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr24_+_29902165 | 0.47 |
ENSDART00000168422
ENSDART00000100092 ENSDART00000113304 |
frrs1b
|
ferric-chelate reductase 1b |
| chr2_-_17443607 | 0.47 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr16_-_31494275 | 0.47 |
ENSDART00000056551
|
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr24_-_29901383 | 0.45 |
ENSDART00000178791
|
CABZ01041804.1
|
ENSDARG00000106630 |
| chr24_+_12845339 | 0.44 |
ENSDART00000126842
|
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr6_-_54340370 | 0.44 |
ENSDART00000156501
|
zgc:101577
|
zgc:101577 |
| chr14_-_26167494 | 0.44 |
ENSDART00000143454
ENSDART00000111860 |
syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr8_+_13753663 | 0.41 |
|
|
|
| chr18_-_19830652 | 0.41 |
ENSDART00000060344
|
aagab
|
alpha- and gamma-adaptin binding protein |
| chr11_-_34909095 | 0.41 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr23_+_30803518 | 0.40 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr10_-_25246786 | 0.40 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr5_+_62611738 | 0.40 |
ENSDART00000177108
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr8_-_1833095 | 0.39 |
ENSDART00000114476
ENSDART00000091235 ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr16_+_41065107 | 0.39 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr5_+_62611606 | 0.38 |
ENSDART00000177108
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr5_-_56810425 | 0.37 |
ENSDART00000147875
ENSDART00000142776 |
ENSDARG00000095415
|
ENSDARG00000095415 |
| chr4_+_4825461 | 0.37 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr15_+_545631 | 0.37 |
ENSDART00000102275
ENSDART00000102274 |
ftr86
|
finTRIM family, member 86 |
| chr15_+_38113499 | 0.37 |
|
|
|
| chr1_+_46639805 | 0.36 |
|
|
|
| chr5_+_62628554 | 0.36 |
ENSDART00000158086
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr23_+_19179865 | 0.36 |
ENSDART00000142429
|
fkbpl
|
FK506 binding protein like |
| chr2_-_31849916 | 0.35 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr4_-_5083532 | 0.35 |
ENSDART00000130453
|
ahcyl2
|
adenosylhomocysteinase-like 2 |
| chr19_+_44073792 | 0.35 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr12_-_33257026 | 0.34 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr18_-_26815261 | 0.34 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr16_-_19636613 | 0.34 |
|
|
|
| chr2_+_43693486 | 0.34 |
|
|
|
| chr23_-_2957262 | 0.33 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr1_-_39177682 | 0.32 |
ENSDART00000035739
|
tmem134
|
transmembrane protein 134 |
| chr16_+_28829383 | 0.32 |
ENSDART00000122433
|
trim33l
|
tripartite motif containing 33, like |
| chr2_+_30497550 | 0.31 |
ENSDART00000125933
|
fam173b
|
family with sequence similarity 173, member B |
| chr2_-_44491143 | 0.31 |
ENSDART00000084174
|
lig1
|
ligase I, DNA, ATP-dependent |
| chr6_-_9471808 | 0.30 |
ENSDART00000039280
|
fzd7b
|
frizzled class receptor 7b |
| chr11_-_24300905 | 0.30 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr23_+_25952913 | 0.29 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr11_-_23458624 | 0.29 |
ENSDART00000124810
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr2_-_58411390 | 0.29 |
ENSDART00000177160
|
AL954696.2
|
ENSDARG00000106834 |
| chr13_-_11873326 | 0.29 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr15_+_1569674 | 0.29 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr6_+_41455676 | 0.29 |
ENSDART00000007798
ENSDART00000162135 |
twf2a
|
twinfilin actin-binding protein 2a |
| chr5_+_62611823 | 0.28 |
ENSDART00000177108
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr23_+_19179748 | 0.28 |
ENSDART00000142429
|
fkbpl
|
FK506 binding protein like |
| KN150266v1_-_68652 | 0.28 |
|
|
|
| chr6_-_40101104 | 0.28 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr5_+_62628471 | 0.28 |
ENSDART00000158086
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr3_-_29837411 | 0.28 |
ENSDART00000139310
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr20_+_9776066 | 0.28 |
ENSDART00000053834
|
psmc6
|
proteasome 26S subunit, ATPase 6 |
| chr1_-_51863300 | 0.28 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr13_+_43266551 | 0.28 |
ENSDART00000084321
|
dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr5_-_64432697 | 0.27 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr9_-_35824470 | 0.27 |
ENSDART00000140356
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr12_+_33257120 | 0.27 |
|
|
|
| chr1_+_46639773 | 0.27 |
|
|
|
| chr15_+_38113014 | 0.26 |
|
|
|
| chr4_-_17033790 | 0.26 |
|
|
|
| chr10_-_8839096 | 0.26 |
ENSDART00000080763
|
si:dkey-27b3.2
|
si:dkey-27b3.2 |
| chr14_+_15698814 | 0.26 |
|
|
|
| chr8_-_24194821 | 0.26 |
ENSDART00000177464
|
CT025875.1
|
ENSDARG00000106046 |
| chr2_-_17443642 | 0.25 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr10_-_35377548 | 0.25 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr23_+_25952724 | 0.25 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr25_+_35540123 | 0.25 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr8_-_14571519 | 0.25 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr3_+_14421976 | 0.24 |
ENSDART00000171731
|
znf653
|
zinc finger protein 653 |
| chr10_-_44713414 | 0.24 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr18_+_2124267 | 0.24 |
|
|
|
| chr2_+_47619337 | 0.24 |
ENSDART00000141974
|
acsl3b
|
acyl-CoA synthetase long-chain family member 3b |
| chr14_+_11803542 | 0.24 |
ENSDART00000121913
|
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr5_+_62611688 | 0.24 |
ENSDART00000177108
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr19_-_32313943 | 0.23 |
ENSDART00000113797
|
zbtb10
|
zinc finger and BTB domain containing 10 |
| chr23_+_25952958 | 0.23 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr23_+_24711233 | 0.23 |
|
|
|
| chr14_-_26167526 | 0.23 |
ENSDART00000143454
ENSDART00000111860 |
syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr1_-_44892558 | 0.23 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr12_-_33256934 | 0.23 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr15_-_545279 | 0.23 |
ENSDART00000155472
|
nudt8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| chr18_-_17735227 | 0.23 |
|
|
|
| KN150702v1_-_111618 | 0.23 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr8_-_25826973 | 0.23 |
ENSDART00000047008
ENSDART00000128829 |
efhd2
|
EF-hand domain family, member D2 |
| chr16_+_34157948 | 0.23 |
ENSDART00000140552
|
tcea3
|
transcription elongation factor A (SII), 3 |
| chr25_-_7526393 | 0.23 |
|
|
|
| chr18_-_26815111 | 0.23 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr19_+_43523515 | 0.23 |
ENSDART00000023156
ENSDART00000039129 |
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr11_-_24300849 | 0.22 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr6_+_23709605 | 0.22 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr16_+_28829508 | 0.22 |
ENSDART00000122433
|
trim33l
|
tripartite motif containing 33, like |
| chr22_+_18904372 | 0.22 |
ENSDART00000131131
|
bsg
|
basigin |
| chr10_-_31861975 | 0.22 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr19_-_10295398 | 0.22 |
ENSDART00000148225
|
znf865
|
zinc finger protein 865 |
| chr10_-_22967737 | 0.22 |
|
|
|
| chr19_+_43523476 | 0.22 |
ENSDART00000023156
ENSDART00000039129 |
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr22_-_10861329 | 0.21 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr19_+_7717962 | 0.21 |
ENSDART00000112404
|
cgnb
|
cingulin b |
| chr3_-_20944652 | 0.21 |
ENSDART00000109790
|
nlk1
|
nemo-like kinase, type 1 |
| chr24_+_37450437 | 0.21 |
ENSDART00000141771
|
gnptg
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit |
| chr21_+_22599022 | 0.21 |
ENSDART00000142495
|
BX510940.1
|
ENSDARG00000089088 |
| chr13_-_402878 | 0.21 |
ENSDART00000038315
|
nvl
|
nuclear VCP-like |
| chr22_+_2244267 | 0.21 |
ENSDART00000142890
|
si:dkey-4c15.9
|
si:dkey-4c15.9 |
| chr7_+_16844277 | 0.20 |
ENSDART00000035558
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr10_+_29964045 | 0.20 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr20_+_27088328 | 0.20 |
ENSDART00000012816
|
sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr15_-_18110169 | 0.20 |
|
|
|
| chr18_-_30526372 | 0.20 |
ENSDART00000028033
|
emc8
|
ER membrane protein complex subunit 8 |
| chr2_-_31850133 | 0.20 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr10_+_21477806 | 0.19 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr3_+_41573382 | 0.19 |
ENSDART00000155440
|
eif3ba
|
eukaryotic translation initiation factor 3, subunit Ba |
| chr7_-_29084775 | 0.19 |
ENSDART00000075757
|
gtf2a2
|
general transcription factor IIA, 2 |
| chr7_-_19116999 | 0.19 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr7_+_34523324 | 0.19 |
ENSDART00000173456
|
esrp2
|
epithelial splicing regulatory protein 2 |
| chr24_-_28253884 | 0.19 |
ENSDART00000149015
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr8_-_14571436 | 0.19 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr11_-_24300785 | 0.18 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr2_+_27730940 | 0.18 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr6_+_41455437 | 0.18 |
ENSDART00000007353
|
wdr82
|
WD repeat domain 82 |
| chr4_+_4825628 | 0.18 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr11_+_34909393 | 0.18 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr3_+_61903372 | 0.18 |
ENSDART00000108945
|
GID4
|
GID complex subunit 4 homolog |
| chr23_+_23256859 | 0.18 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr21_-_22321244 | 0.18 |
ENSDART00000136904
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr23_-_2957169 | 0.18 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr5_+_23559435 | 0.17 |
ENSDART00000142268
|
gps2
|
G protein pathway suppressor 2 |
| chr23_-_19905270 | 0.17 |
ENSDART00000054479
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr25_+_18855868 | 0.17 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr13_-_24130160 | 0.17 |
ENSDART00000138747
ENSDART00000101168 |
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr24_-_36307897 | 0.17 |
ENSDART00000154395
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr16_-_41585555 | 0.17 |
ENSDART00000131706
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr13_-_24265471 | 0.17 |
ENSDART00000016211
|
tbp
|
TATA box binding protein |
| chr20_-_33594156 | 0.17 |
ENSDART00000101974
|
erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr15_-_14653696 | 0.16 |
ENSDART00000172195
|
adck4
|
aarF domain containing kinase 4 |
| chr9_+_30822402 | 0.16 |
|
|
|
| chr15_+_20428823 | 0.16 |
ENSDART00000062676
|
supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
| chr10_+_3481047 | 0.16 |
ENSDART00000092684
|
rph3aa
|
rabphilin 3A homolog (mouse), a |
| chr23_-_3813451 | 0.15 |
|
|
|
| chr8_+_46319434 | 0.15 |
ENSDART00000145618
|
si:dkey-75a21.2
|
si:dkey-75a21.2 |
| chr5_-_12815350 | 0.15 |
ENSDART00000143364
|
sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr3_-_52406525 | 0.15 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr20_-_25745974 | 0.15 |
ENSDART00000063137
ENSDART00000082099 |
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr13_+_9852854 | 0.15 |
ENSDART00000022051
|
gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr15_-_545179 | 0.15 |
ENSDART00000155472
|
nudt8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| chr10_+_34057791 | 0.15 |
ENSDART00000149934
|
kl
|
klotho |
| chr6_-_58980624 | 0.15 |
ENSDART00000144911
|
mars
|
methionyl-tRNA synthetase |
| chr15_-_8786639 | 0.15 |
ENSDART00000154171
|
arhgap35a
|
Rho GTPase activating protein 35a |
| KN150529v1_+_2610 | 0.14 |
|
|
|
| chr6_-_8125287 | 0.14 |
ENSDART00000139161
ENSDART00000140021 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr15_+_6462447 | 0.14 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr6_-_39220951 | 0.14 |
ENSDART00000133305
ENSDART00000166290 |
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr23_-_2957203 | 0.14 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr9_+_3547661 | 0.14 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| KN149858v1_+_13014 | 0.14 |
ENSDART00000165916
|
MED8
|
mediator complex subunit 8 |
| chr19_+_42658184 | 0.14 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr10_+_22921629 | 0.14 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr7_-_19116856 | 0.14 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr12_+_27151693 | 0.14 |
ENSDART00000066269
|
arl4d
|
ADP-ribosylation factor-like 4D |
| chr15_-_38113404 | 0.13 |
|
|
|
| chr3_-_36277628 | 0.13 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr19_+_44073707 | 0.13 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr8_-_14516930 | 0.13 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr1_-_51863187 | 0.13 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr18_+_48868656 | 0.13 |
|
|
|
| chr18_-_510340 | 0.13 |
ENSDART00000164374
|
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr19_+_42657913 | 0.13 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr2_-_52862423 | 0.13 |
|
|
|
| chr5_-_21430034 | 0.13 |
|
|
|
| chr13_-_402908 | 0.13 |
ENSDART00000038315
|
nvl
|
nuclear VCP-like |
| chr23_-_39367876 | 0.13 |
|
|
|
| chr16_+_4288700 | 0.13 |
ENSDART00000159694
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr22_-_10128498 | 0.13 |
ENSDART00000152042
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr22_-_37092134 | 0.12 |
|
|
|
| chr7_+_28814408 | 0.12 |
ENSDART00000173960
|
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr21_-_37750338 | 0.12 |
ENSDART00000133585
|
fam114a2
|
family with sequence similarity 114, member A2 |
| chr20_+_4000402 | 0.12 |
ENSDART00000112053
|
fam89a
|
family with sequence similarity 89, member A |
| chr5_+_57784991 | 0.12 |
ENSDART00000038602
|
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.8 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 1.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.5 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.6 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 0.5 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.3 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.9 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.7 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.8 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.2 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) interferon-gamma-mediated signaling pathway(GO:0060333) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 1.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0043393 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 1.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.0 | 1.2 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.2 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:1903513 | endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 0.2 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.1 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.2 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.0 | 0.8 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.1 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.2 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.1 | GO:0010517 | regulation of phospholipase activity(GO:0010517) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.4 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.1 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.6 | GO:0006414 | translational elongation(GO:0006414) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.7 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.9 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 0.7 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.3 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.6 | GO:0008199 | acid phosphatase activity(GO:0003993) ferric iron binding(GO:0008199) |
| 0.1 | 0.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0008665 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0003948 | N4-(beta-N-acetylglucosaminyl)-L-asparaginase activity(GO:0003948) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |