Project
DANIO-CODE
Navigation
Downloads
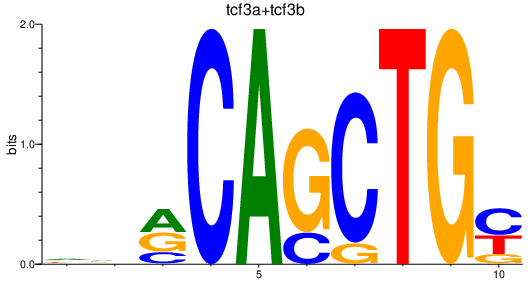
Results for tcf3a+tcf3b
Z-value: 0.89
Transcription factors associated with tcf3a+tcf3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf3a
|
ENSDARG00000005915 | transcription factor 3a |
|
tcf3b
|
ENSDARG00000099999 | transcription factor 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf3a | dr10_dc_chr2_-_57244608_57244772 | 0.67 | 4.7e-03 | Click! |
| tcf3b | dr10_dc_chr22_-_20316908_20317043 | 0.64 | 7.2e-03 | Click! |
Activity profile of tcf3a+tcf3b motif
Sorted Z-values of tcf3a+tcf3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf3a+tcf3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_23711689 | 2.67 |
ENSDART00000128644
|
ckmb
|
creatine kinase, muscle b |
| chr2_-_16896965 | 2.11 |
ENSDART00000022549
|
atp1b3a
|
ATPase, Na+/K+ transporting, beta 3a polypeptide |
| chr11_-_5868257 | 2.09 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr2_-_21693984 | 1.96 |
ENSDART00000171699
|
hhatla
|
hedgehog acyltransferase-like, a |
| chr15_+_34131126 | 1.83 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr14_+_11151485 | 1.77 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr24_+_17125429 | 1.68 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr22_+_26543146 | 1.68 |
|
|
|
| chr21_+_27346176 | 1.61 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr4_+_8637319 | 1.61 |
|
|
|
| chr19_+_38834972 | 1.60 |
ENSDART00000140198
ENSDART00000033037 ENSDART00000035093 |
col9a2
|
procollagen, type IX, alpha 2 |
| chr11_-_30105047 | 1.53 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr6_-_42006033 | 1.50 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr16_+_3090170 | 1.48 |
ENSDART00000110395
|
limd1a
|
LIM domains containing 1a |
| chr22_+_11726312 | 1.46 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr8_-_44305494 | 1.44 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr16_+_24057260 | 1.43 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr16_+_49796978 | 1.42 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr19_+_38834765 | 1.42 |
ENSDART00000140198
ENSDART00000033037 ENSDART00000035093 |
col9a2
|
procollagen, type IX, alpha 2 |
| chr6_-_39315024 | 1.40 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr14_+_21531709 | 1.39 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr1_-_50066633 | 1.38 |
|
|
|
| chr7_+_6517248 | 1.36 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr8_-_985673 | 1.35 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr17_+_27417635 | 1.34 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr12_-_5085227 | 1.34 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr4_-_14330113 | 1.34 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr16_-_9978112 | 1.33 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr16_+_737615 | 1.33 |
ENSDART00000161774
|
irx1a
|
iroquois homeobox 1a |
| chr23_-_7892862 | 1.32 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr14_-_25659535 | 1.31 |
ENSDART00000135627
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr4_+_12618256 | 1.29 |
ENSDART00000112860
ENSDART00000134362 |
lmo3
|
LIM domain only 3 |
| chr5_-_36237656 | 1.27 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr16_+_5625552 | 1.27 |
|
|
|
| chr24_-_33817169 | 1.25 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr22_+_21997000 | 1.25 |
ENSDART00000046174
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| KN150178v1_-_23644 | 1.24 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr16_-_29229687 | 1.24 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr4_+_12618485 | 1.24 |
ENSDART00000112860
|
lmo3
|
LIM domain only 3 |
| chr15_+_7190427 | 1.23 |
ENSDART00000109394
|
her13
|
hairy-related 13 |
| chr24_+_35676505 | 1.20 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr10_+_16634200 | 1.17 |
ENSDART00000101142
|
chsy3
|
chondroitin sulfate synthase 3 |
| chr15_-_15421065 | 1.17 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr23_+_43362722 | 1.16 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr16_+_46327528 | 1.14 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr1_+_51925783 | 1.12 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr11_-_25803101 | 1.11 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr14_-_36523075 | 1.11 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr10_-_10372266 | 1.09 |
|
|
|
| chr1_-_40208469 | 1.08 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr23_-_10747605 | 1.08 |
|
|
|
| chr17_+_8166167 | 1.07 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr11_-_41357639 | 1.06 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr20_+_4567653 | 1.06 |
|
|
|
| chr1_-_54319943 | 1.06 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr8_-_11191783 | 1.06 |
ENSDART00000131171
|
unc45b
|
unc-45 myosin chaperone B |
| chr18_-_6494014 | 1.05 |
ENSDART00000062423
|
tnni1c
|
troponin I, skeletal, slow c |
| chr9_+_42019770 | 1.05 |
ENSDART00000097295
|
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr8_-_7049946 | 1.05 |
ENSDART00000045669
ENSDART00000162950 |
KRT73
|
keratin 73 |
| chr18_-_3244618 | 1.04 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr21_+_549788 | 1.04 |
ENSDART00000175116
|
CABZ01100207.1
|
ENSDARG00000106163 |
| chr13_-_31310439 | 1.04 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr19_+_31046291 | 1.04 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr12_-_36138709 | 1.03 |
ENSDART00000130985
|
st6galnac
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase |
| chr19_-_23037220 | 1.03 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr7_+_25861386 | 1.03 |
ENSDART00000129834
|
nat16
|
N-acetyltransferase 16 |
| chr17_-_6219019 | 1.02 |
|
|
|
| chr19_-_22182031 | 1.02 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr2_+_55859099 | 1.02 |
ENSDART00000097753
ENSDART00000141688 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr24_-_3334678 | 1.01 |
|
|
|
| chr9_-_23081250 | 1.01 |
|
|
|
| chr18_+_38307946 | 1.01 |
ENSDART00000134247
|
lmo2
|
LIM domain only 2 (rhombotin-like 1) |
| chr6_+_3519697 | 1.00 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| KN149932v1_+_27584 | 1.00 |
|
|
|
| chr10_+_15819127 | 0.99 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr2_-_42279180 | 0.99 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr14_+_5852295 | 0.98 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr6_-_39053400 | 0.98 |
ENSDART00000165839
|
tns2b
|
tensin 2b |
| chr9_+_3458086 | 0.98 |
ENSDART00000160977
ENSDART00000114168 |
CU469503.1
itga6a
|
ENSDARG00000099348 integrin, alpha 6a |
| chr6_-_19432410 | 0.97 |
ENSDART00000006843
|
cacng1a
|
calcium channel, voltage-dependent, gamma subunit 1a |
| chr22_-_14103803 | 0.97 |
ENSDART00000062902
|
ENSDARG00000042857
|
ENSDARG00000042857 |
| chr23_-_10242377 | 0.96 |
ENSDART00000129044
|
krt5
|
keratin 5 |
| chr22_-_613948 | 0.94 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr22_+_11745592 | 0.94 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr19_-_2011177 | 0.93 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr8_+_25748377 | 0.93 |
ENSDART00000062406
|
cacna1sb
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, b |
| chr6_+_4712785 | 0.93 |
ENSDART00000151674
|
pcdh9
|
protocadherin 9 |
| chr7_+_40972616 | 0.93 |
ENSDART00000173814
|
scrib
|
scribbled planar cell polarity protein |
| chr5_-_25866099 | 0.92 |
ENSDART00000144035
|
arvcfb
|
armadillo repeat gene deleted in velocardiofacial syndrome b |
| chr17_+_6119323 | 0.91 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr7_+_23778570 | 0.91 |
ENSDART00000121837
|
efs
|
embryonal Fyn-associated substrate |
| chr12_+_22734974 | 0.91 |
ENSDART00000130594
|
afap1
|
actin filament associated protein 1 |
| chr15_+_29343644 | 0.91 |
ENSDART00000170537
ENSDART00000128973 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr9_-_23081918 | 0.89 |
ENSDART00000143888
|
neb
|
nebulin |
| chr16_-_14184394 | 0.89 |
ENSDART00000090234
|
trim109
|
tripartite motif containing 109 |
| chr8_+_54171992 | 0.89 |
ENSDART00000122692
|
CABZ01112317.1
|
ENSDARG00000086057 |
| chr23_-_9924987 | 0.88 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr16_+_6923898 | 0.88 |
ENSDART00000078306
|
arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr2_-_44402486 | 0.87 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr8_-_19167286 | 0.87 |
ENSDART00000161646
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr24_+_20414578 | 0.87 |
ENSDART00000131677
|
hhatlb
|
hedgehog acyltransferase-like, b |
| chr19_-_7531709 | 0.86 |
ENSDART00000104750
|
mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr14_-_41311458 | 0.86 |
ENSDART00000163039
|
fgfrl1b
|
fibroblast growth factor receptor-like 1b |
| chr7_+_58718614 | 0.85 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr16_+_1073570 | 0.85 |
|
|
|
| chr9_-_4890648 | 0.84 |
ENSDART00000167998
|
fmnl2a
|
formin-like 2a |
| chr18_-_54695 | 0.84 |
|
|
|
| chr22_-_16009772 | 0.83 |
|
|
|
| chr1_-_16894589 | 0.83 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long-chain family member 1a |
| chr11_-_32461160 | 0.83 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr3_+_17387551 | 0.82 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr24_+_7832020 | 0.82 |
ENSDART00000019705
|
bmp6
|
bone morphogenetic protein 6 |
| chr10_+_26705729 | 0.82 |
ENSDART00000147013
|
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr12_+_27032862 | 0.82 |
|
|
|
| chr24_+_4341242 | 0.82 |
ENSDART00000133360
|
ccny
|
cyclin Y |
| chr23_-_18945009 | 0.82 |
ENSDART00000080064
|
CR847953.1
|
ENSDARG00000057403 |
| chr8_+_15987710 | 0.82 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr23_+_36832325 | 0.82 |
|
|
|
| chr10_+_42530040 | 0.82 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr5_-_40133824 | 0.82 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr13_-_3022033 | 0.82 |
ENSDART00000050934
|
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr10_+_34377959 | 0.81 |
|
|
|
| chr9_+_3147601 | 0.81 |
ENSDART00000135619
|
ftr52p
|
finTRIM family, member 52, pseudogene |
| chr4_-_802415 | 0.81 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr2_-_32372801 | 0.80 |
ENSDART00000143348
|
BX663499.1
|
ENSDARG00000093778 |
| chr23_-_3466041 | 0.80 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr6_-_42006225 | 0.80 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr3_-_1463509 | 0.79 |
ENSDART00000053892
|
ENSDARG00000074266
|
ENSDARG00000074266 |
| chr22_-_14103942 | 0.79 |
ENSDART00000140323
|
ENSDARG00000042857
|
ENSDARG00000042857 |
| chr7_+_31567166 | 0.79 |
ENSDART00000099785
ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr17_+_53068731 | 0.79 |
ENSDART00000156774
|
dph6
|
diphthamine biosynthesis 6 |
| chr3_+_60637167 | 0.78 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr15_-_19789174 | 0.78 |
ENSDART00000152345
ENSDART00000047643 ENSDART00000163435 |
sytl2b
|
synaptotagmin-like 2b |
| chr22_+_38096040 | 0.78 |
ENSDART00000097533
|
wwtr1
|
WW domain containing transcription regulator 1 |
| chr23_-_15807415 | 0.78 |
ENSDART00000178557
|
BX005011.3
|
ENSDARG00000108083 |
| chr25_-_2386464 | 0.78 |
|
|
|
| chr15_+_28435937 | 0.77 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr4_-_4825948 | 0.77 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr24_+_5205878 | 0.77 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr5_-_37851313 | 0.77 |
ENSDART00000136428
ENSDART00000112487 |
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr22_+_24131239 | 0.77 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr12_+_29994735 | 0.77 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr9_+_23854859 | 0.76 |
ENSDART00000060905
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr7_+_49442972 | 0.76 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr6_-_15526547 | 0.76 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr18_+_38773971 | 0.76 |
ENSDART00000010177
|
onecut1
|
one cut homeobox 1 |
| chr15_-_18226612 | 0.76 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr23_+_25782195 | 0.76 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr13_+_22733062 | 0.76 |
ENSDART00000113082
|
hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr20_-_26568204 | 0.75 |
ENSDART00000158213
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr22_-_17652938 | 0.75 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr5_+_36180981 | 0.75 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr21_-_25486257 | 0.75 |
ENSDART00000110923
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr22_+_12327849 | 0.75 |
ENSDART00000178678
|
FO704572.1
|
ENSDARG00000106890 |
| chr8_-_40231417 | 0.75 |
ENSDART00000162020
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr14_-_8975187 | 0.75 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr23_-_31585883 | 0.75 |
ENSDART00000157511
|
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr1_+_38834751 | 0.75 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr3_+_36373519 | 0.75 |
ENSDART00000161652
|
si:dkeyp-72e1.9
|
si:dkeyp-72e1.9 |
| chr6_-_12487617 | 0.74 |
ENSDART00000090174
|
dock9b
|
dedicator of cytokinesis 9b |
| chr22_+_11745657 | 0.74 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr18_+_30391910 | 0.74 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
| chr22_+_4722336 | 0.73 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr11_-_25592102 | 0.73 |
ENSDART00000103549
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr6_-_9964616 | 0.73 |
|
|
|
| chr18_-_11216447 | 0.73 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr24_+_38267919 | 0.73 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr10_+_34372205 | 0.72 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr4_-_1399909 | 0.72 |
|
|
|
| chr11_-_11317608 | 0.72 |
ENSDART00000113311
|
col9a1a
|
collagen, type IX, alpha 1a |
| chr11_+_666006 | 0.71 |
ENSDART00000130839
|
timp4.1
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 1 |
| chr22_+_16282153 | 0.71 |
ENSDART00000162685
ENSDART00000105678 |
lrrc39
|
leucine rich repeat containing 39 |
| chr24_+_26287471 | 0.71 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr9_+_1502716 | 0.71 |
ENSDART00000137230
ENSDART00000093427 |
pde11a
|
phosphodiesterase 11a |
| chr5_+_64411412 | 0.71 |
ENSDART00000158484
|
BX001023.2
|
ENSDARG00000101163 |
| chr8_+_39964695 | 0.71 |
ENSDART00000073782
|
ggt5a
|
gamma-glutamyltransferase 5a |
| chr1_-_674449 | 0.71 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr9_-_49834232 | 0.71 |
|
|
|
| chr18_+_9413588 | 0.71 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr9_+_41720118 | 0.71 |
|
|
|
| chr7_+_13753344 | 0.71 |
|
|
|
| chr9_+_30774173 | 0.70 |
ENSDART00000160590
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr19_-_2939818 | 0.70 |
|
|
|
| chr4_+_4841191 | 0.70 |
ENSDART00000130818
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr21_-_4842528 | 0.70 |
ENSDART00000097796
|
rnf165a
|
ring finger protein 165a |
| chr5_-_18895882 | 0.70 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr12_-_25825072 | 0.70 |
|
|
|
| chr10_-_23939618 | 0.69 |
|
|
|
| chr8_+_46319434 | 0.69 |
ENSDART00000145618
|
si:dkey-75a21.2
|
si:dkey-75a21.2 |
| chr24_+_4946079 | 0.69 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr17_+_45430353 | 0.69 |
ENSDART00000162937
|
ezra
|
ezrin a |
| chr1_+_33563354 | 0.69 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr5_-_21520111 | 0.68 |
|
|
|
| chr15_+_9351511 | 0.68 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr16_-_26663643 | 0.68 |
ENSDART00000008152
|
sgk2b
|
serum/glucocorticoid regulated kinase 2b |
| chr11_+_24720057 | 0.68 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr14_+_21999898 | 0.68 |
|
|
|
| chr12_+_16845837 | 0.67 |
ENSDART00000138174
|
slc16a12b
|
solute carrier family 16, member 12b |
| chr6_-_19132923 | 0.67 |
|
|
|
| chr20_-_32429820 | 0.66 |
ENSDART00000062982
|
foxo3b
|
forkhead box O3b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:1903060 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.7 | 2.1 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.6 | 1.7 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.5 | 3.9 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.4 | 1.3 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.4 | 1.2 | GO:0061381 | associative learning(GO:0008306) visual learning(GO:0008542) mammillary axonal complex development(GO:0061373) mammillothalamic axonal tract development(GO:0061374) corpora quadrigemina development(GO:0061378) inferior colliculus development(GO:0061379) cell migration in diencephalon(GO:0061381) |
| 0.3 | 1.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 1.0 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.3 | 1.0 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.3 | 0.9 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.3 | 0.9 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.3 | GO:0060259 | regulation of behavior(GO:0050795) regulation of feeding behavior(GO:0060259) |
| 0.3 | 2.6 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.3 | 1.1 | GO:0032374 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.3 | 0.8 | GO:0030431 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.3 | 1.3 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.3 | 0.8 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.3 | 0.8 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.3 | 0.5 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 0.7 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 2.7 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 1.0 | GO:1901389 | transforming growth factor beta activation(GO:0036363) transforming growth factor beta production(GO:0071604) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.2 | 0.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 0.7 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.2 | 1.4 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.2 | 0.9 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 0.9 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 0.6 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 1.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 0.8 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 2.0 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 0.6 | GO:0097186 | enamel mineralization(GO:0070166) amelogenesis(GO:0097186) |
| 0.2 | 0.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 0.4 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.2 | 1.9 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.2 | 1.1 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 0.8 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.2 | 0.9 | GO:0045639 | megakaryocyte differentiation(GO:0030219) positive regulation of myeloid cell differentiation(GO:0045639) |
| 0.2 | 0.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.2 | 0.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.1 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.2 | 1.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 1.3 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.2 | 0.7 | GO:0050687 | negative regulation of response to biotic stimulus(GO:0002832) negative regulation of defense response to virus(GO:0050687) |
| 0.2 | 0.8 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.2 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.9 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.1 | 0.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 2.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 2.1 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.7 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 1.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.0 | GO:0060579 | amacrine cell differentiation(GO:0035881) ventral spinal cord interneuron fate commitment(GO:0060579) |
| 0.1 | 0.8 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.7 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.9 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.7 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.5 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.8 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 1.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.4 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 1.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.8 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.7 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.1 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.1 | 0.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.6 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.3 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.4 | GO:1901741 | positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.9 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 1.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.8 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.4 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 0.4 | GO:1901381 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 2.3 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 0.5 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.2 | GO:2000136 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.1 | 0.4 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 0.3 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.1 | 1.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.6 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.1 | 0.8 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.4 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.4 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.4 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.5 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.1 | 0.3 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 1.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.8 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.5 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.3 | GO:2000379 | protein import into mitochondrial inner membrane(GO:0045039) positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.1 | 1.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.4 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 1.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 1.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.2 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 0.2 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.7 | GO:0051149 | positive regulation of myoblast differentiation(GO:0045663) positive regulation of muscle cell differentiation(GO:0051149) |
| 0.1 | 0.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 1.2 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 0.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.2 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.1 | 1.1 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 0.3 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.1 | 1.0 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 0.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.5 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.2 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 1.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0098921 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.6 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 1.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.3 | GO:0035909 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.5 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:1902414 | protein localization to adherens junction(GO:0071896) protein localization to cell junction(GO:1902414) |
| 0.0 | 4.2 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.8 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.5 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 1.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 1.5 | GO:0018210 | peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 2.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.6 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 1.3 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.3 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.0 | 0.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 2.0 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 1.5 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 2.4 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.3 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.5 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.5 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.8 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 0.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.7 | GO:0051896 | protein kinase B signaling(GO:0043491) regulation of protein kinase B signaling(GO:0051896) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 1.4 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.3 | GO:0060606 | neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) tube closure(GO:0060606) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.3 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0047496 | vesicle transport along microtubule(GO:0047496) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.3 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.5 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.5 | GO:0051693 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.0 | 0.4 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.4 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.8 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.1 | GO:0070142 | synaptic vesicle budding(GO:0070142) |
| 0.0 | 0.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.0 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.2 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.0 | 0.8 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.3 | 0.8 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.2 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 2.1 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 1.1 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:0008352 | katanin complex(GO:0008352) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 2.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 5.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 2.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 2.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.4 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.0 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0098594 | mucin granule(GO:0098594) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.2 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.0 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 3.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 11.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.9 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 2.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 0.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 0.9 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.3 | 1.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 0.7 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 1.7 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.2 | 1.0 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.2 | 1.1 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 1.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 1.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 1.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.5 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 0.6 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.4 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 1.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.7 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 1.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 2.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.8 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.3 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.1 | 1.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.7 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.1 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 5.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.8 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.5 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.4 | GO:0035381 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 2.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.5 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.4 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.8 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 1.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 1.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0016894 | endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 3'-phosphomonoesters(GO:0016894) |
| 0.0 | 1.3 | GO:0045499 | semaphorin receptor binding(GO:0030215) chemorepellent activity(GO:0045499) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 1.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 1.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.5 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.6 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.5 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 4.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 4.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 23.3 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0034979 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 0.9 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.7 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 2.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 2.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.8 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.2 | 1.3 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.2 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 3.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 1.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |