Project
DANIO-CODE
Navigation
Downloads
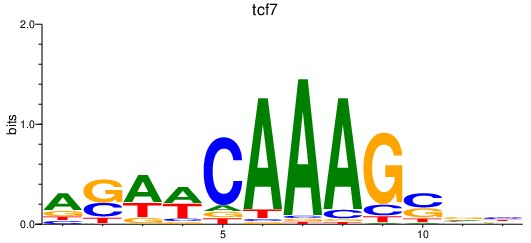
Results for tcf7
Z-value: 0.75
Transcription factors associated with tcf7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf7
|
ENSDARG00000038672 | transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf7 | dr10_dc_chr21_+_45227200_45227294 | -0.71 | 2.0e-03 | Click! |
Activity profile of tcf7 motif
Sorted Z-values of tcf7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_540624 | 2.77 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr17_+_23463072 | 1.88 |
|
|
|
| chr1_-_7160294 | 1.70 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr14_+_1226767 | 1.41 |
ENSDART00000127477
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr1_-_7160099 | 1.40 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr20_+_39145226 | 1.30 |
ENSDART00000142778
|
BX569784.1
|
ENSDARG00000092980 |
| chr1_-_40671573 | 1.24 |
ENSDART00000074777
ENSDART00000158114 |
htt
|
huntingtin |
| chr1_+_23685562 | 1.23 |
ENSDART00000102542
ENSDART00000160882 |
ENSDARG00000090822
qdprb2
|
ENSDARG00000090822 quinoid dihydropteridine reductase b2 |
| chr12_+_48356793 | 1.23 |
ENSDART00000054788
ENSDART00000152899 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr17_-_14583728 | 1.20 |
ENSDART00000174703
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr24_+_25872005 | 1.19 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr5_-_27549500 | 1.17 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr1_+_47323244 | 1.10 |
|
|
|
| chr19_-_7114660 | 1.07 |
ENSDART00000124094
|
daxx
|
death-domain associated protein |
| chr1_+_47323043 | 1.01 |
|
|
|
| chr3_-_26052785 | 0.99 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr5_-_41396159 | 0.99 |
ENSDART00000143573
|
ncor1
|
nuclear receptor corepressor 1 |
| chr20_-_31594202 | 0.97 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr14_-_6637670 | 0.97 |
ENSDART00000126279
|
clk4a
|
CDC-like kinase 4a |
| chr14_-_36101782 | 0.94 |
|
|
|
| chr7_-_73613531 | 0.91 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr23_+_540664 | 0.91 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr21_-_18238558 | 0.90 |
ENSDART00000126672
|
brd3a
|
bromodomain containing 3a |
| chr21_+_15339960 | 0.90 |
ENSDART00000137491
|
si:dkey-11o15.10
|
si:dkey-11o15.10 |
| KN149710v1_+_38638 | 0.84 |
|
|
|
| chr5_-_23192997 | 0.83 |
ENSDART00000167629
|
rnf128a
|
ring finger protein 128a |
| chr24_+_25872150 | 0.83 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr21_+_13291305 | 0.81 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr19_+_43468369 | 0.80 |
ENSDART00000165202
|
pum1
|
pumilio RNA-binding family member 1 |
| chr3_-_59690168 | 0.79 |
ENSDART00000035878
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr7_-_23840878 | 0.78 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr1_-_7159912 | 0.77 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr21_+_13291242 | 0.76 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr5_-_3323004 | 0.75 |
|
|
|
| chr5_-_23192934 | 0.74 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr3_-_60394005 | 0.74 |
ENSDART00000053502
|
DXO
|
decapping exoribonuclease |
| chr2_-_30800968 | 0.72 |
ENSDART00000056735
|
rgs20
|
regulator of G protein signaling 20 |
| chr25_-_35820568 | 0.72 |
ENSDART00000152766
|
CR354435.4
|
Histone H2B 1/2 |
| chr5_-_56294194 | 0.72 |
ENSDART00000074400
|
tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr11_-_23458624 | 0.71 |
ENSDART00000124810
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr7_-_22361808 | 0.71 |
ENSDART00000159867
ENSDART00000165706 |
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr3_-_26060047 | 0.70 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr15_+_47207251 | 0.70 |
ENSDART00000154481
|
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr2_+_21516908 | 0.69 |
ENSDART00000079518
|
dnajc1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr3_-_29760552 | 0.68 |
ENSDART00000014021
|
slc25a39
|
solute carrier family 25, member 39 |
| chr3_-_29760674 | 0.68 |
ENSDART00000014021
|
slc25a39
|
solute carrier family 25, member 39 |
| chr12_+_48295188 | 0.67 |
ENSDART00000164427
|
ndr2
|
nodal-related 2 |
| chr10_-_32716555 | 0.66 |
ENSDART00000063544
|
atg101
|
autophagy related 101 |
| chr22_+_11943005 | 0.65 |
ENSDART00000105788
|
mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr3_-_26052601 | 0.65 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr7_-_23841075 | 0.64 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr2_-_27892824 | 0.62 |
|
|
|
| chr10_+_16543577 | 0.61 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr8_+_26015350 | 0.60 |
ENSDART00000004521
|
arih2
|
ariadne homolog 2 (Drosophila) |
| chr23_-_39253291 | 0.59 |
|
|
|
| chr24_-_5904097 | 0.58 |
ENSDART00000007373
ENSDART00000135124 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr9_-_36065830 | 0.57 |
ENSDART00000013432
|
dnajc28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr21_+_13291347 | 0.57 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr15_-_36072087 | 0.57 |
ENSDART00000076229
|
irs1
|
insulin receptor substrate 1 |
| chr21_+_18238176 | 0.56 |
ENSDART00000144322
|
wdr5
|
WD repeat domain 5 |
| chr8_+_26015379 | 0.54 |
ENSDART00000142555
|
arih2
|
ariadne homolog 2 (Drosophila) |
| chr2_+_1645259 | 0.54 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr14_+_6122803 | 0.53 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr23_-_20050732 | 0.50 |
ENSDART00000054659
|
fam58a
|
family with sequence similarity 58, member A |
| chr7_-_57206977 | 0.50 |
ENSDART00000147036
|
sirt3
|
sirtuin 3 |
| chr14_-_23811519 | 0.50 |
ENSDART00000124255
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr1_-_40671522 | 0.50 |
ENSDART00000074777
ENSDART00000158114 |
htt
|
huntingtin |
| chr7_-_54407355 | 0.49 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr2_+_36957471 | 0.48 |
ENSDART00000143915
|
gadd45bb
|
growth arrest and DNA-damage-inducible, beta b |
| chr23_+_116397 | 0.47 |
|
|
|
| chr5_-_23727210 | 0.46 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr16_-_12206435 | 0.46 |
ENSDART00000103944
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr6_-_6818607 | 0.46 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr11_-_3313199 | 0.44 |
ENSDART00000002545
|
mcrs1
|
microspherule protein 1 |
| chr5_+_66693263 | 0.44 |
ENSDART00000142156
|
sebox
|
SEBOX homeobox |
| chr22_-_10511906 | 0.44 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr3_-_60394265 | 0.43 |
ENSDART00000053502
|
DXO
|
decapping exoribonuclease |
| chr14_-_23980060 | 0.42 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr17_-_20675997 | 0.42 |
ENSDART00000063613
ENSDART00000171575 |
ccdc6a
|
coiled-coil domain containing 6a |
| chr7_-_22361743 | 0.42 |
ENSDART00000159867
ENSDART00000165706 |
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr15_+_47207100 | 0.42 |
ENSDART00000154481
|
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr17_+_23463668 | 0.40 |
|
|
|
| chr7_-_6297068 | 0.39 |
ENSDART00000172954
|
si:ch1073-153i20.4
|
si:ch1073-153i20.4 |
| chr12_+_30728864 | 0.39 |
|
|
|
| chr4_-_3015097 | 0.38 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr4_-_3014778 | 0.38 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr16_+_25222407 | 0.38 |
ENSDART00000177098
|
znf576.1
|
zinc finger protein 576, tandem duplicate 1 |
| chr3_-_1377802 | 0.38 |
ENSDART00000149061
|
si:ch73-211l13.2
|
si:ch73-211l13.2 |
| chr25_+_4508802 | 0.38 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr12_+_26971687 | 0.38 |
|
|
|
| chr5_-_22981395 | 0.37 |
ENSDART00000170293
|
si:dkeyp-20g2.1
|
si:dkeyp-20g2.1 |
| chr7_+_7463565 | 0.36 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr22_-_20236913 | 0.36 |
ENSDART00000134485
|
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr7_-_22361507 | 0.36 |
ENSDART00000165245
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr15_-_23849212 | 0.35 |
ENSDART00000059354
|
rad1
|
RAD1 homolog (S. pombe) |
| chr23_-_20050588 | 0.34 |
ENSDART00000054659
|
fam58a
|
family with sequence similarity 58, member A |
| chr23_+_1693691 | 0.34 |
ENSDART00000149357
ENSDART00000099024 |
rabggta
|
Rab geranylgeranyltransferase, alpha subunit |
| chr10_+_16544000 | 0.33 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr12_-_11311554 | 0.32 |
ENSDART00000079645
|
zgc:174164
|
zgc:174164 |
| chr9_+_46944755 | 0.31 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr17_-_8828004 | 0.31 |
|
|
|
| chr20_+_4000402 | 0.31 |
ENSDART00000112053
|
fam89a
|
family with sequence similarity 89, member A |
| chr12_+_48295296 | 0.30 |
ENSDART00000164427
|
ndr2
|
nodal-related 2 |
| chr24_-_5903682 | 0.30 |
ENSDART00000007373
ENSDART00000135124 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr10_-_33399226 | 0.30 |
ENSDART00000164191
|
c2cd2
|
C2 calcium-dependent domain containing 2 |
| chr11_+_26366532 | 0.28 |
ENSDART00000159505
|
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr20_+_28367799 | 0.28 |
ENSDART00000153319
ENSDART00000103330 |
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr16_+_3170784 | 0.28 |
|
|
|
| chr1_-_52155939 | 0.27 |
ENSDART00000139577
|
patl1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr3_+_7127701 | 0.26 |
|
|
|
| chr20_+_39319873 | 0.26 |
ENSDART00000019744
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr10_-_32716794 | 0.25 |
ENSDART00000141013
|
atg101
|
autophagy related 101 |
| chr22_-_800449 | 0.24 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr18_+_44635321 | 0.24 |
|
|
|
| chr21_-_40759658 | 0.24 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr1_+_31608918 | 0.23 |
ENSDART00000137156
|
pudp
|
pseudouridine 5'-phosphatase |
| chr5_-_22981367 | 0.23 |
ENSDART00000134069
|
si:dkeyp-20g2.3
|
si:dkeyp-20g2.3 |
| chr5_-_23727236 | 0.22 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr14_+_6655894 | 0.22 |
ENSDART00000148394
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr4_-_71046688 | 0.22 |
ENSDART00000161800
|
CABZ01054391.1
|
ENSDARG00000104152 |
| chr4_+_75469488 | 0.21 |
ENSDART00000144892
|
znf1009
|
zinc finger protein 1009 |
| chr10_+_29963102 | 0.20 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr12_-_29118423 | 0.20 |
ENSDART00000153175
|
si:ch211-214e3.5
|
si:ch211-214e3.5 |
| chr24_+_37911828 | 0.20 |
ENSDART00000131975
|
telo2
|
TEL2, telomere maintenance 2, homolog (S. cerevisiae) |
| chr19_-_43468824 | 0.20 |
|
|
|
| chr15_-_23550450 | 0.19 |
ENSDART00000152706
|
nlrx1
|
NLR family member X1 |
| chr14_+_6655541 | 0.19 |
ENSDART00000148447
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr20_+_31375056 | 0.18 |
|
|
|
| chr18_-_6394175 | 0.18 |
ENSDART00000009217
|
ddx11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr1_+_599358 | 0.16 |
ENSDART00000135944
|
jam2a
|
junctional adhesion molecule 2a |
| chr17_-_21037758 | 0.16 |
ENSDART00000048853
|
ube2d1a
|
ubiquitin-conjugating enzyme E2D 1a |
| chr25_-_3168287 | 0.16 |
|
|
|
| chr12_+_26971596 | 0.15 |
|
|
|
| chr1_-_58536658 | 0.14 |
ENSDART00000163021
|
mvb12a
|
multivesicular body subunit 12A |
| chr4_-_747097 | 0.13 |
ENSDART00000103601
|
agbl5
|
ATP/GTP binding protein-like 5 |
| chr25_+_3168376 | 0.12 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr2_+_47769055 | 0.12 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr1_-_40671600 | 0.10 |
ENSDART00000074777
ENSDART00000158114 |
htt
|
huntingtin |
| chr1_-_49650100 | 0.10 |
ENSDART00000053028
|
tbck
|
TBC1 domain containing kinase |
| chr1_+_264808 | 0.09 |
ENSDART00000139438
|
cenpe
|
centromere protein E |
| chr3_-_23513293 | 0.08 |
ENSDART00000140264
|
eve1
|
even-skipped-like1 |
| chr7_+_73601671 | 0.08 |
ENSDART00000163075
|
zgc:173552
|
zgc:173552 |
| chr23_-_29897606 | 0.08 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr19_+_43468063 | 0.08 |
ENSDART00000168263
|
pum1
|
pumilio RNA-binding family member 1 |
| chr12_+_26972001 | 0.08 |
|
|
|
| chr23_+_27125377 | 0.08 |
ENSDART00000054237
|
ptges3a
|
prostaglandin E synthase 3a (cytosolic) |
| chr21_-_40759739 | 0.07 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr9_+_46944989 | 0.07 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr8_-_1201388 | 0.07 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr14_-_23980261 | 0.07 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr7_+_20266209 | 0.06 |
ENSDART00000052916
|
ENSDARG00000036426
|
ENSDARG00000036426 |
| chr24_+_25872051 | 0.06 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr14_+_6656015 | 0.05 |
ENSDART00000148394
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr1_-_54028333 | 0.05 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr12_+_6432058 | 0.04 |
ENSDART00000066477
|
dkk1b
|
dickkopf WNT signaling pathway inhibitor 1b |
| chr21_-_41191143 | 0.04 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr8_-_1200407 | 0.03 |
ENSDART00000148654
|
cdc14b
|
cell division cycle 14B |
| chr24_-_11767965 | 0.03 |
ENSDART00000033621
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr21_+_21246921 | 0.03 |
|
|
|
| chr1_-_17922028 | 0.03 |
ENSDART00000139550
|
klf3
|
Kruppel-like factor 3 (basic) |
| chr1_-_33694144 | 0.02 |
|
|
|
| chr22_+_10630419 | 0.02 |
ENSDART00000105835
|
tusc2b
|
tumor suppressor candidate 2b |
| chr20_-_52133991 | 0.02 |
ENSDART00000074325
|
dusp10
|
dual specificity phosphatase 10 |
| chr2_-_23331500 | 0.01 |
ENSDART00000146497
|
mllt1b
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 1b |
| chr25_-_12691936 | 0.01 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr17_-_10684072 | 0.01 |
|
|
|
| chr9_+_13762263 | 0.01 |
ENSDART00000141314
ENSDART00000138254 |
abi2a
|
abl-interactor 2a |
| chr9_-_137883 | 0.00 |
ENSDART00000169377
|
FQ377903.3
|
ENSDARG00000104933 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0031548 | neural plate formation(GO:0021990) brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) anterior neural plate formation(GO:0090017) |
| 0.4 | 2.1 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.3 | 1.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 1.2 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 0.6 | GO:0070657 | mechanosensory epithelium regeneration(GO:0070655) mechanoreceptor differentiation involved in mechanosensory epithelium regeneration(GO:0070656) neuromast regeneration(GO:0070657) neuromast hair cell differentiation involved in neuromast regeneration(GO:0070658) |
| 0.2 | 0.6 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.2 | 1.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 1.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 1.0 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 0.4 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.9 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.5 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.9 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 1.2 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 1.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:1902592 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.0 | 0.2 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.0 | 0.7 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.9 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.6 | GO:0036294 | cellular response to decreased oxygen levels(GO:0036294) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.0 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.0 | 0.9 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.7 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 1.9 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.3 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.2 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 2.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.7 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 1.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.4 | 1.5 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 1.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 1.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 1.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.4 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 3.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 1.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 3.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |