Project
DANIO-CODE
Navigation
Downloads
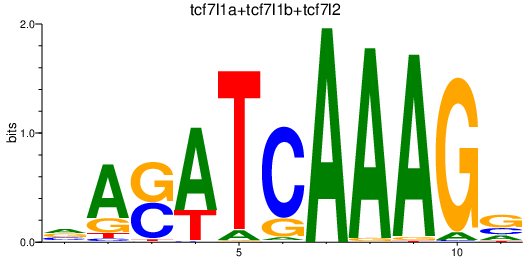
Results for tcf7l1a+tcf7l1b+tcf7l2
Z-value: 1.48
Transcription factors associated with tcf7l1a+tcf7l1b+tcf7l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf7l2
|
ENSDARG00000004415 | transcription factor 7 like 2 |
|
tcf7l1b
|
ENSDARG00000007369 | transcription factor 7 like 1b |
|
tcf7l1a
|
ENSDARG00000038159 | transcription factor 7 like 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf7l2 | dr10_dc_chr12_-_30988279_30988485 | 0.90 | 1.9e-06 | Click! |
| tcf7l1a | dr10_dc_chr10_-_42469083_42469113 | 0.76 | 6.3e-04 | Click! |
| tcf7l1b | dr10_dc_chr8_-_52243483_52243655 | 0.73 | 1.5e-03 | Click! |
Activity profile of tcf7l1a+tcf7l1b+tcf7l2 motif
Sorted Z-values of tcf7l1a+tcf7l1b+tcf7l2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf7l1a+tcf7l1b+tcf7l2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_45950530 | 6.84 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr5_+_41680352 | 5.79 |
ENSDART00000142855
|
tbx6l
|
T-box 6, like |
| chr7_-_54407681 | 4.31 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr15_-_35563367 | 3.81 |
|
|
|
| chr5_-_23829861 | 3.70 |
ENSDART00000051553
|
znf703
|
zinc finger protein 703 |
| chr25_-_22089794 | 3.59 |
ENSDART00000139110
|
pkp3a
|
plakophilin 3a |
| chr1_+_16683931 | 3.45 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr14_+_23980348 | 3.27 |
ENSDART00000160984
|
BX005228.1
|
ENSDARG00000105465 |
| chr3_+_54876680 | 3.12 |
ENSDART00000111585
|
hbbe1.3
|
hemoglobin, beta embryonic 1.3 |
| chr12_-_30988279 | 3.08 |
ENSDART00000122972
ENSDART00000005562 ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 |
tcf7l2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr17_-_20877606 | 3.08 |
ENSDART00000088106
|
ank3a
|
ankyrin 3a |
| chr25_+_21732255 | 3.07 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr9_-_9693163 | 2.92 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr21_-_41286846 | 2.83 |
ENSDART00000167339
|
msx2b
|
muscle segment homeobox 2b |
| chr7_-_54407461 | 2.82 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr16_+_32605735 | 2.64 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr23_-_22186674 | 2.62 |
ENSDART00000142474
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr8_-_41194452 | 2.57 |
ENSDART00000165949
|
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr3_+_26896869 | 2.53 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr1_+_25662652 | 2.50 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr4_+_292818 | 2.45 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr10_+_32160464 | 2.43 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr1_+_46537108 | 2.42 |
|
|
|
| chr13_+_7575753 | 2.38 |
|
|
|
| chr21_+_45227200 | 2.37 |
ENSDART00000155681
ENSDART00000157136 |
tcf7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr1_-_35419335 | 2.36 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr11_-_22966274 | 2.34 |
ENSDART00000003646
|
optc
|
opticin |
| chr3_+_39425125 | 2.34 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr15_-_11257445 | 2.30 |
|
|
|
| chr9_+_44233139 | 2.29 |
ENSDART00000162406
|
itga4
|
integrin alpha 4 |
| chr18_+_45199548 | 2.27 |
|
|
|
| chr7_-_72500748 | 2.22 |
ENSDART00000160523
|
CU929444.1
|
ENSDARG00000099109 |
| chr23_-_2084541 | 2.21 |
ENSDART00000091532
|
ndnf
|
neuron-derived neurotrophic factor |
| chr3_-_23513177 | 2.14 |
ENSDART00000078425
|
eve1
|
even-skipped-like1 |
| chr7_+_38479571 | 2.10 |
ENSDART00000170486
|
f2
|
coagulation factor II (thrombin) |
| chr6_+_30441419 | 2.09 |
|
|
|
| chr13_-_50299821 | 2.08 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr22_+_21221777 | 2.07 |
ENSDART00000079046
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr21_-_20291707 | 1.99 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr22_-_16015935 | 1.97 |
ENSDART00000062633
|
s1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr7_-_37350985 | 1.90 |
ENSDART00000173523
|
nkd1
|
naked cuticle homolog 1 (Drosophila) |
| chr19_-_9603597 | 1.90 |
ENSDART00000045245
|
ing4
|
inhibitor of growth family, member 4 |
| chr23_-_26150495 | 1.87 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr4_-_2187291 | 1.80 |
ENSDART00000150490
|
ENSDARG00000009262
|
ENSDARG00000009262 |
| chr16_-_8481813 | 1.73 |
ENSDART00000051166
|
grb10b
|
growth factor receptor-bound protein 10b |
| chr9_+_17298410 | 1.70 |
ENSDART00000048548
|
scel
|
sciellin |
| chr20_-_40553998 | 1.67 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr16_+_26900732 | 1.63 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr13_-_50299643 | 1.63 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr4_-_17420378 | 1.62 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr12_+_27444832 | 1.57 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr16_-_32605560 | 1.55 |
|
|
|
| chr19_+_30210743 | 1.53 |
ENSDART00000167803
|
marcksl1b
|
MARCKS-like 1b |
| chr8_+_39525254 | 1.52 |
|
|
|
| chr13_+_4312466 | 1.50 |
ENSDART00000156323
|
tb
|
T, brachyury homolog b |
| chr13_+_28601627 | 1.49 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr20_+_27194162 | 1.49 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr9_-_3700395 | 1.48 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr14_+_22150325 | 1.47 |
ENSDART00000108585
ENSDART00000161776 |
sowahab
|
sosondowah ankyrin repeat domain family member Ab |
| chr8_-_51767046 | 1.47 |
ENSDART00000007090
|
tbx16
|
T-box 16 |
| chr14_+_34154895 | 1.46 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr6_+_58921655 | 1.45 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr13_-_31310439 | 1.45 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr1_-_40208544 | 1.42 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr19_+_42899678 | 1.40 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr21_-_25486257 | 1.31 |
ENSDART00000110923
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr15_-_8333117 | 1.29 |
ENSDART00000061351
|
tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr5_-_28970034 | 1.28 |
ENSDART00000043259
|
entpd2a.2
|
ectonucleoside triphosphate diphosphohydrolase 2a, tandem duplicate 2 |
| chr23_-_20837809 | 1.25 |
ENSDART00000089750
|
znf362b
|
zinc finger protein 362b |
| chr7_-_18302231 | 1.25 |
ENSDART00000108938
|
msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr20_+_25326042 | 1.24 |
ENSDART00000130494
|
moxd1
|
monooxygenase, DBH-like 1 |
| chr6_-_2294751 | 1.23 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr1_-_28241500 | 1.23 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr6_-_54103765 | 1.23 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr13_-_18704185 | 1.20 |
ENSDART00000146795
|
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr6_-_43806125 | 1.19 |
|
|
|
| chr7_-_9557990 | 1.17 |
ENSDART00000055593
|
aldh1a3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr6_-_26569321 | 1.17 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr7_+_69418011 | 1.17 |
ENSDART00000166105
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr8_-_41194530 | 1.16 |
ENSDART00000173055
|
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr1_-_40208469 | 1.15 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr15_-_2224784 | 1.15 |
ENSDART00000009564
|
shox2
|
short stature homeobox 2 |
| chr8_-_18868986 | 1.15 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr14_-_23980261 | 1.15 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr8_-_30233470 | 1.12 |
ENSDART00000139864
|
zgc:162939
|
zgc:162939 |
| chr5_+_18827478 | 1.11 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr24_-_23155932 | 1.11 |
|
|
|
| chr14_+_34151963 | 1.10 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr12_+_6432058 | 1.07 |
ENSDART00000066477
|
dkk1b
|
dickkopf WNT signaling pathway inhibitor 1b |
| chr12_-_31342432 | 1.07 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr15_-_2969420 | 1.05 |
|
|
|
| chr2_-_59293269 | 1.04 |
ENSDART00000148286
|
ftr39p
|
finTRIM family, member 39, pseudogene |
| chr20_+_22766887 | 1.03 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr3_-_55399331 | 1.03 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr7_+_37105966 | 1.01 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr22_-_29886668 | 1.01 |
ENSDART00000125017
|
BX649294.1
|
ENSDARG00000092881 |
| chr20_+_23599157 | 0.91 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr20_-_35173530 | 0.90 |
ENSDART00000019196
|
lft1
|
lefty1 |
| chr20_+_23392782 | 0.90 |
|
|
|
| chr20_-_48677794 | 0.88 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr19_+_43523303 | 0.87 |
ENSDART00000167847
|
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr13_-_31516399 | 0.84 |
ENSDART00000172375
ENSDART00000125987 ENSDART00000143903 ENSDART00000158719 |
six4a
|
SIX homeobox 4a |
| chr20_+_22766918 | 0.83 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr7_+_73594790 | 0.83 |
ENSDART00000123081
|
zgc:173552
|
zgc:173552 |
| chr3_-_34417082 | 0.83 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr24_+_9124068 | 0.82 |
|
|
|
| chr17_+_23102259 | 0.81 |
ENSDART00000178403
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr18_-_39602605 | 0.78 |
ENSDART00000125116
|
tnfaip8l3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr19_+_43524098 | 0.77 |
ENSDART00000129362
|
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr16_+_24046789 | 0.76 |
|
|
|
| chr1_+_26418707 | 0.73 |
ENSDART00000161169
|
bnc2
|
basonuclin 2 |
| chr10_-_31619761 | 0.72 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr1_+_51192660 | 0.70 |
ENSDART00000052992
|
dand5
|
DAN domain family, member 5 |
| chr2_-_53797472 | 0.70 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr6_+_15635696 | 0.69 |
ENSDART00000128939
|
iqca1
|
IQ motif containing with AAA domain 1 |
| chr13_-_40027211 | 0.69 |
|
|
|
| chr24_+_11194381 | 0.68 |
ENSDART00000143171
|
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr14_-_25154966 | 0.68 |
ENSDART00000148652
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr11_-_19436304 | 0.65 |
ENSDART00000110499
|
atxn7
|
ataxin 7 |
| chr16_+_5411816 | 0.65 |
|
|
|
| chr22_-_29886840 | 0.64 |
ENSDART00000125017
|
BX649294.1
|
ENSDARG00000092881 |
| chr12_+_23717554 | 0.63 |
ENSDART00000152997
|
svila
|
supervillin a |
| chr18_-_14868444 | 0.63 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr21_+_44203036 | 0.63 |
|
|
|
| chr13_+_32987105 | 0.63 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr5_+_37113743 | 0.61 |
|
|
|
| chr13_+_33138144 | 0.61 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr5_-_41645167 | 0.60 |
|
|
|
| chr1_+_46537333 | 0.60 |
|
|
|
| chr3_-_55399433 | 0.58 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr6_-_6818925 | 0.58 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr13_+_26943081 | 0.58 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr5_+_26127909 | 0.58 |
ENSDART00000098545
|
tmem150aa
|
transmembrane protein 150Aa |
| chr4_+_17088146 | 0.57 |
|
|
|
| chr6_+_15635738 | 0.57 |
ENSDART00000128939
|
iqca1
|
IQ motif containing with AAA domain 1 |
| chr20_-_9448803 | 0.56 |
ENSDART00000133000
|
ENSDARG00000033201
|
ENSDARG00000033201 |
| chr14_+_6655894 | 0.56 |
ENSDART00000148394
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr20_+_41652149 | 0.56 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr1_-_35419273 | 0.54 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr24_-_35486496 | 0.54 |
|
|
|
| chr5_-_38873781 | 0.54 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr1_-_42078442 | 0.52 |
ENSDART00000074618
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr1_-_7744605 | 0.52 |
ENSDART00000033917
|
syngr3b
|
synaptogyrin 3b |
| chr19_+_43468063 | 0.52 |
ENSDART00000168263
|
pum1
|
pumilio RNA-binding family member 1 |
| chr21_-_25486330 | 0.51 |
ENSDART00000162711
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr13_-_31516518 | 0.49 |
ENSDART00000172375
ENSDART00000125987 ENSDART00000143903 ENSDART00000158719 |
six4a
|
SIX homeobox 4a |
| chr1_-_52024906 | 0.48 |
ENSDART00000150568
|
aptx
|
aprataxin |
| chr23_-_40059749 | 0.48 |
ENSDART00000158168
|
CU657980.1
|
ENSDARG00000054846 |
| chr3_+_28700619 | 0.48 |
ENSDART00000108973
|
flr
|
fleer |
| chr24_+_32635139 | 0.47 |
ENSDART00000175986
|
CU138525.1
|
ENSDARG00000105889 |
| chr6_-_11544518 | 0.47 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr19_+_43414160 | 0.47 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr3_-_28297082 | 0.46 |
ENSDART00000151546
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr3_-_36940675 | 0.45 |
ENSDART00000026701
|
tubg1
|
tubulin, gamma 1 |
| chr2_-_43234323 | 0.45 |
ENSDART00000084303
|
kcnq3
|
potassium voltage-gated channel, KQT-like subfamily, member 3 |
| chr4_+_16721264 | 0.45 |
ENSDART00000035899
|
pkp2
|
plakophilin 2 |
| chr2_-_53797503 | 0.45 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr17_+_26593564 | 0.45 |
ENSDART00000159811
|
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr22_-_22696091 | 0.45 |
|
|
|
| chr11_-_39940309 | 0.43 |
ENSDART00000086287
|
znf362a
|
zinc finger protein 362a |
| chr22_+_21221950 | 0.41 |
ENSDART00000079046
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr5_-_21578927 | 0.40 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr21_+_26576104 | 0.39 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr6_+_51676410 | 0.39 |
ENSDART00000151565
|
fam65c
|
family with sequence similarity 65, member C |
| chr6_-_2294717 | 0.39 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr18_+_36825511 | 0.38 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr2_-_30005139 | 0.38 |
ENSDART00000146968
|
cnpy1
|
canopy1 |
| KN150672v1_+_74532 | 0.37 |
|
|
|
| chr5_-_7591260 | 0.37 |
ENSDART00000158447
|
nipbla
|
nipped-B homolog a (Drosophila) |
| chr3_-_23513293 | 0.36 |
ENSDART00000140264
|
eve1
|
even-skipped-like1 |
| chr9_+_7570199 | 0.34 |
ENSDART00000081543
|
ptprna
|
protein tyrosine phosphatase, receptor type, Na |
| chr14_-_31748733 | 0.34 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr14_+_20862466 | 0.34 |
ENSDART00000059796
|
ENSDARG00000019213
|
ENSDARG00000019213 |
| chr20_-_54364559 | 0.34 |
|
|
|
| chr13_+_7575720 | 0.32 |
|
|
|
| chr6_-_54103833 | 0.31 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr11_-_41487157 | 0.31 |
ENSDART00000173312
|
abhd6a
|
abhydrolase domain containing 6a |
| chr9_+_50341397 | 0.31 |
|
|
|
| chr15_+_28173909 | 0.30 |
ENSDART00000041707
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr9_+_30183691 | 0.29 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr14_+_6656015 | 0.27 |
ENSDART00000148394
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr12_+_27444762 | 0.26 |
ENSDART00000013033
|
etv4
|
ets variant 4 |
| chr1_-_58577305 | 0.26 |
|
|
|
| chr23_+_26090659 | 0.26 |
ENSDART00000167427
ENSDART00000002939 |
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr23_+_26090616 | 0.25 |
ENSDART00000167427
ENSDART00000002939 |
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr3_-_2010512 | 0.24 |
|
|
|
| chr25_+_22474108 | 0.24 |
ENSDART00000161559
|
stra6
|
stimulated by retinoic acid 6 |
| chr9_+_7570161 | 0.24 |
ENSDART00000081543
|
ptprna
|
protein tyrosine phosphatase, receptor type, Na |
| chr21_+_19797435 | 0.23 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr14_-_38532216 | 0.22 |
ENSDART00000127479
|
gsr
|
glutathione reductase |
| chr13_+_4040744 | 0.22 |
|
|
|
| chr3_-_26847908 | 0.22 |
ENSDART00000155396
ENSDART00000077817 |
nubp1
|
nucleotide binding protein 1 (MinD homolog, E. coli) |
| chr12_+_27444732 | 0.21 |
ENSDART00000013033
|
etv4
|
ets variant 4 |
| chr14_+_6655541 | 0.20 |
ENSDART00000148447
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr8_-_12292501 | 0.20 |
|
|
|
| chr18_-_8828500 | 0.18 |
|
|
|
| chr9_-_28588288 | 0.18 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr21_+_19797559 | 0.18 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr3_+_41573049 | 0.17 |
ENSDART00000155440
|
eif3ba
|
eukaryotic translation initiation factor 3, subunit Ba |
| chr14_+_14759351 | 0.17 |
ENSDART00000167075
|
nkx1.2lb
|
NK1 transcription factor related 2-like,b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 1.3 | 6.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic G1 DNA damage checkpoint(GO:0031571) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.8 | 3.1 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.7 | 2.2 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.6 | 2.6 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.6 | 6.2 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.6 | 3.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.5 | 2.1 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.5 | 2.5 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.5 | 2.0 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.5 | 1.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.5 | 1.9 | GO:1903533 | regulation of protein targeting(GO:1903533) |
| 0.5 | 2.4 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.4 | 1.3 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.4 | 1.2 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.4 | 1.2 | GO:0001504 | neurotransmitter uptake(GO:0001504) regulation of neurotransmitter uptake(GO:0051580) neurotransmitter reuptake(GO:0098810) |
| 0.4 | 1.6 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.4 | 3.1 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.4 | 1.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.4 | 1.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.4 | 3.7 | GO:0034333 | adherens junction assembly(GO:0034333) |
| 0.4 | 1.1 | GO:0003197 | endocardial cushion development(GO:0003197) hepatic duct development(GO:0061011) |
| 0.4 | 1.1 | GO:2000193 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 2.5 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.3 | 1.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.3 | 1.7 | GO:0035545 | determination of left/right asymmetry in diencephalon(GO:0035462) determination of left/right asymmetry in nervous system(GO:0035545) |
| 0.3 | 3.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.7 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 6.8 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.2 | 3.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.3 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.2 | 0.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.9 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 1.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 2.3 | GO:0030388 | thigmotaxis(GO:0001966) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.8 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.1 | 0.7 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.5 | GO:0036372 | opsin transport(GO:0036372) |
| 0.1 | 0.6 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 2.6 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 4.0 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.1 | 0.7 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 2.3 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.6 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 1.5 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 0.2 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.7 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 2.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 2.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.2 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.2 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.0 | 0.4 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 1.7 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.6 | 2.6 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.4 | 3.1 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 3.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 3.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 3.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 7.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 0.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 1.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 2.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.4 | GO:0044304 | main axon(GO:0044304) |
| 0.1 | 2.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 3.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.8 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.5 | 2.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.5 | 2.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.4 | 3.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 2.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.4 | 3.1 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 1.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 1.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 1.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 2.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 1.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 3.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 5.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 6.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 2.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.2 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 2.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 26.5 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.7 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 2.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 2.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 2.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.2 | 2.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 6.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.0 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |