Project
DANIO-CODE
Navigation
Downloads
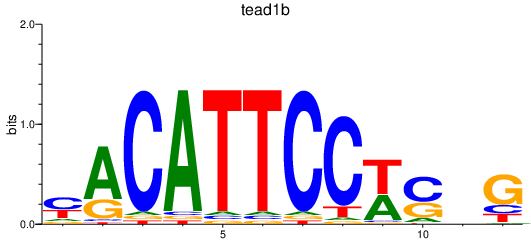
Results for tead1b_tead1a
Z-value: 1.23
Transcription factors associated with tead1b_tead1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tead1b
|
ENSDARG00000059483 | TEA domain family member 1b |
|
tead1a
|
ENSDARG00000028159 | TEA domain family member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tead1a | dr10_dc_chr25_+_16259634_16259838 | -0.43 | 9.7e-02 | Click! |
Activity profile of tead1b_tead1a motif
Sorted Z-values of tead1b_tead1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tead1b_tead1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_53353690 | 4.49 |
ENSDART00000126339
|
dla
|
deltaA |
| chr8_-_38126693 | 4.14 |
ENSDART00000112331
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr13_+_1595986 | 4.09 |
|
|
|
| chr13_-_46292762 | 3.83 |
ENSDART00000149125
|
fgfr2
|
fibroblast growth factor receptor 2 |
| chr23_+_35984675 | 3.73 |
ENSDART00000053295
|
hoxc10a
|
homeobox C10a |
| chr5_-_43334777 | 3.43 |
ENSDART00000142271
|
ENSDARG00000053091
|
ENSDARG00000053091 |
| chr15_+_5935952 | 3.22 |
ENSDART00000152520
ENSDART00000145827 |
sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr21_+_26660833 | 3.09 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| KN150131v1_+_59098 | 3.01 |
ENSDART00000175590
|
CABZ01064941.1
|
ENSDARG00000106550 |
| chr3_-_16387246 | 2.92 |
ENSDART00000133907
|
eps8l1
|
eps8-like1 |
| chr18_-_605558 | 2.91 |
ENSDART00000157564
|
wtip
|
WT1 interacting protein |
| chr7_-_3900223 | 2.73 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr20_-_29148810 | 2.47 |
ENSDART00000140350
|
thbs1b
|
thrombospondin 1b |
| chr1_-_37695900 | 2.45 |
ENSDART00000018533
|
hpgd
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr20_+_25441689 | 2.45 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr25_+_22756060 | 2.43 |
ENSDART00000073588
|
kcnj11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr23_+_25428372 | 2.42 |
ENSDART00000147440
|
fmnl3
|
formin-like 3 |
| chr5_-_68270398 | 2.32 |
ENSDART00000161561
|
ENSDARG00000059093
|
ENSDARG00000059093 |
| chr13_-_46292975 | 2.29 |
ENSDART00000167402
ENSDART00000080914 ENSDART00000080916 |
fgfr2
|
fibroblast growth factor receptor 2 |
| chr5_-_70948223 | 2.16 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr1_-_18709814 | 2.13 |
ENSDART00000145224
|
rbm47
|
RNA binding motif protein 47 |
| chr7_-_44332679 | 2.11 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr5_-_31708933 | 2.07 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr13_+_1398289 | 2.01 |
|
|
|
| chr15_-_20088439 | 1.95 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr23_+_10552781 | 1.94 |
|
|
|
| chr14_-_20766448 | 1.92 |
ENSDART00000164146
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr6_+_7092350 | 1.89 |
ENSDART00000049695
ENSDART00000136088 ENSDART00000083424 ENSDART00000125912 |
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr6_+_29420644 | 1.88 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr17_+_38619041 | 1.83 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr7_-_23474701 | 1.81 |
ENSDART00000048050
|
ITGB1BP2
|
integrin subunit beta 1 binding protein 2 |
| chr23_-_30119058 | 1.80 |
ENSDART00000103480
|
ccdc187
|
coiled-coil domain containing 187 |
| chr17_+_32619043 | 1.80 |
ENSDART00000087565
|
eva1a
|
eva-1 homolog A (C. elegans) |
| chr9_-_23406315 | 1.73 |
ENSDART00000159256
|
kif5c
|
kinesin family member 5C |
| chr2_+_31399487 | 1.72 |
ENSDART00000086863
|
colec12
|
collectin sub-family member 12 |
| chr23_+_35984885 | 1.71 |
ENSDART00000130260
|
hoxc10a
|
homeobox C10a |
| KN150670v1_-_32201 | 1.71 |
|
|
|
| chr17_-_20938261 | 1.66 |
ENSDART00000113655
|
tmem26a
|
transmembrane protein 26a |
| chr12_+_22536231 | 1.64 |
ENSDART00000163755
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr7_-_44332885 | 1.56 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr13_-_46292862 | 1.53 |
ENSDART00000150061
|
fgfr2
|
fibroblast growth factor receptor 2 |
| chr2_-_844040 | 1.52 |
ENSDART00000028159
|
foxf2a
|
forkhead box F2a |
| chr23_-_17543731 | 1.51 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr6_+_7309023 | 1.48 |
ENSDART00000160128
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr23_+_27725660 | 1.47 |
|
|
|
| chr9_-_12604260 | 1.47 |
ENSDART00000102419
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr7_-_71566426 | 1.44 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr23_+_20522512 | 1.42 |
ENSDART00000137294
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr1_-_58258428 | 1.40 |
|
|
|
| chr21_-_36495488 | 1.39 |
|
|
|
| chr16_+_21109486 | 1.34 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr14_+_45895103 | 1.33 |
|
|
|
| chr15_-_18130992 | 1.32 |
ENSDART00000113142
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr19_-_5851328 | 1.29 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr5_+_43365449 | 1.27 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr6_-_49512935 | 1.27 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr9_-_23222916 | 1.25 |
ENSDART00000022392
ENSDART00000122036 |
rnd3b
|
Rho family GTPase 3b |
| chr22_-_18087170 | 1.23 |
|
|
|
| chr7_-_21978889 | 1.21 |
|
|
|
| chr6_+_57744081 | 1.20 |
ENSDART00000169175
|
SNTA1
|
syntrophin alpha 1 |
| chr7_-_37587227 | 1.18 |
ENSDART00000173552
|
adcy7
|
adenylate cyclase 7 |
| chr2_-_44330405 | 1.18 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr1_+_15880988 | 1.17 |
ENSDART00000166317
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr15_+_21399127 | 1.16 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr9_+_30483319 | 1.16 |
ENSDART00000148148
|
srpx
|
sushi-repeat containing protein, X-linked |
| chr11_+_743048 | 1.14 |
ENSDART00000172972
|
vgll4b
|
vestigial-like family member 4b |
| chr12_+_27370834 | 1.13 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr3_-_16387615 | 1.12 |
ENSDART00000080749
ENSDART00000133824 |
eps8l1
|
eps8-like1 |
| chr15_+_21399299 | 1.10 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr7_-_3900148 | 1.10 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr14_+_34440699 | 1.09 |
ENSDART00000130469
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr18_+_12179227 | 1.08 |
ENSDART00000162067
ENSDART00000168386 |
fgd4a
|
FYVE, RhoGEF and PH domain containing 4a |
| chr5_+_58446427 | 1.07 |
ENSDART00000154978
|
si:ch211-284f22.3
|
si:ch211-284f22.3 |
| chr14_-_29552242 | 1.06 |
ENSDART00000088004
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr14_-_45883839 | 1.06 |
|
|
|
| chr15_+_29823641 | 1.01 |
|
|
|
| chr11_-_25592102 | 0.99 |
ENSDART00000103549
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr23_-_2027166 | 0.98 |
ENSDART00000127443
|
prdm5
|
PR domain containing 5 |
| chr14_-_29459799 | 0.96 |
ENSDART00000005568
|
pdlim3b
|
PDZ and LIM domain 3b |
| chr9_-_34691159 | 0.95 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr8_+_40604188 | 0.94 |
|
|
|
| chr3_-_30833046 | 0.94 |
|
|
|
| chr8_-_18502159 | 0.93 |
ENSDART00000148802
ENSDART00000149081 ENSDART00000148962 |
nexn
|
nexilin (F actin binding protein) |
| chr9_-_34690993 | 0.92 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr4_-_824333 | 0.92 |
ENSDART00000135618
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr3_-_39208943 | 0.91 |
|
|
|
| chr17_+_1058137 | 0.89 |
|
|
|
| chr11_-_25592017 | 0.88 |
ENSDART00000103549
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr15_+_21399228 | 0.88 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr21_+_10699071 | 0.83 |
|
|
|
| chr13_-_10539949 | 0.82 |
ENSDART00000132393
|
slc3a1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr24_+_11194381 | 0.82 |
ENSDART00000143171
|
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr9_-_34691081 | 0.81 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr9_-_12604530 | 0.81 |
ENSDART00000128931
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr14_+_28136958 | 0.80 |
|
|
|
| chr14_+_41805073 | 0.79 |
ENSDART00000074362
|
pcdh18b
|
protocadherin 18b |
| chr9_+_52126790 | 0.78 |
|
|
|
| chr20_-_25727145 | 0.77 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr20_-_25726868 | 0.77 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr13_+_26943081 | 0.76 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr20_+_46683031 | 0.76 |
ENSDART00000153231
|
jdp2b
|
Jun dimerization protein 2b |
| chr19_-_44044039 | 0.76 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr25_-_13456748 | 0.75 |
ENSDART00000139290
|
ano10b
|
anoctamin 10b |
| chr17_-_31041977 | 0.74 |
ENSDART00000104307
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr14_+_41804998 | 0.73 |
ENSDART00000074362
|
pcdh18b
|
protocadherin 18b |
| chr14_-_46407410 | 0.73 |
ENSDART00000173078
|
si:ch211-168f7.5
|
si:ch211-168f7.5 |
| chr17_-_8481455 | 0.73 |
ENSDART00000148971
|
ctbp2a
|
C-terminal binding protein 2a |
| chr19_+_19916407 | 0.72 |
ENSDART00000166755
|
tril
|
TLR4 interactor with leucine-rich repeats |
| chr5_-_51184375 | 0.71 |
ENSDART00000163616
|
homer1b
|
homer scaffolding protein 1b |
| KN150670v1_-_31884 | 0.71 |
|
|
|
| chr14_+_29951389 | 0.71 |
ENSDART00000173004
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr23_-_27930478 | 0.71 |
ENSDART00000043462
|
acvrl1
|
activin A receptor type II-like 1 |
| chr4_+_7499654 | 0.70 |
ENSDART00000158999
|
tnnt2e
|
troponin T2e, cardiac |
| chr1_-_30546026 | 0.70 |
ENSDART00000085370
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
| chr7_+_16615075 | 0.70 |
|
|
|
| chr16_-_29342511 | 0.70 |
ENSDART00000058870
|
rhbg
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr8_-_15192275 | 0.69 |
ENSDART00000141763
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr14_+_4044357 | 0.68 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr9_+_26292183 | 0.68 |
|
|
|
| chr15_-_4490770 | 0.67 |
ENSDART00000156930
|
tfdp2
|
transcription factor Dp-2 |
| chr17_-_50140718 | 0.65 |
ENSDART00000058707
|
jdp2a
|
Jun dimerization protein 2a |
| chr13_+_15084591 | 0.65 |
ENSDART00000133041
|
pank2
|
pantothenate kinase 2 |
| chr8_+_31863748 | 0.64 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr15_-_2882099 | 0.64 |
|
|
|
| chr11_+_38873704 | 0.63 |
|
|
|
| chr21_+_10698503 | 0.63 |
ENSDART00000144460
|
znf532
|
zinc finger protein 532 |
| chr18_+_27926238 | 0.63 |
ENSDART00000142336
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr13_-_49695322 | 0.62 |
|
|
|
| chr9_+_50341397 | 0.62 |
|
|
|
| chr23_+_18796386 | 0.60 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr6_-_39009073 | 0.59 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr10_-_38206238 | 0.58 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr7_-_3900346 | 0.58 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr19_+_40661722 | 0.58 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr1_-_43323814 | 0.58 |
ENSDART00000148932
|
si:ch73-109d9.1
|
si:ch73-109d9.1 |
| chr16_+_45772666 | 0.56 |
ENSDART00000154704
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr7_-_55214437 | 0.53 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr24_+_34720673 | 0.53 |
ENSDART00000105477
|
lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr12_+_31558667 | 0.52 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr5_-_3077874 | 0.51 |
|
|
|
| chr9_+_7745593 | 0.50 |
ENSDART00000061716
|
mnx2a
|
motor neuron and pancreas homeobox 2a |
| chr15_-_3120500 | 0.50 |
|
|
|
| chr19_+_5399813 | 0.50 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr22_-_18087242 | 0.46 |
|
|
|
| chr20_+_20831085 | 0.46 |
ENSDART00000047662
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr25_-_20886847 | 0.45 |
ENSDART00000110879
|
magi2b
|
membrane associated guanylate kinase, WW and PDZ domain containing 2b |
| chr16_+_31890795 | 0.45 |
ENSDART00000159821
|
atn1
|
atrophin 1 |
| chr19_+_5399877 | 0.43 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr4_-_21745163 | 0.42 |
ENSDART00000139962
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr14_-_30467807 | 0.42 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr3_-_41369324 | 0.41 |
|
|
|
| chr2_+_21976053 | 0.41 |
ENSDART00000089822
|
fbxl7
|
F-box and leucine-rich repeat protein 7 |
| chr24_+_17056023 | 0.39 |
ENSDART00000149225
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr23_-_27930393 | 0.38 |
ENSDART00000043462
|
acvrl1
|
activin A receptor type II-like 1 |
| chr5_+_69383382 | 0.37 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr14_-_20766762 | 0.37 |
ENSDART00000164373
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr15_-_12297979 | 0.37 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr10_-_38206060 | 0.37 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr6_+_32428877 | 0.35 |
|
|
|
| chr25_-_11287103 | 0.34 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr24_-_33370476 | 0.34 |
ENSDART00000155429
|
zgc:195173
|
zgc:195173 |
| chr6_+_38882803 | 0.34 |
ENSDART00000132798
|
bin2b
|
bridging integrator 2b |
| chr14_+_14541725 | 0.34 |
ENSDART00000169932
|
ccdc142
|
coiled-coil domain containing 142 |
| chr6_-_45903830 | 0.33 |
ENSDART00000025428
|
epha2a
|
eph receptor A2 a |
| chr24_-_25021505 | 0.33 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr21_-_36495244 | 0.33 |
|
|
|
| chr9_+_42805274 | 0.28 |
ENSDART00000138133
|
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr22_-_20141216 | 0.28 |
ENSDART00000128023
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr15_+_6462607 | 0.28 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr10_-_38206132 | 0.28 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr11_-_11722452 | 0.27 |
ENSDART00000161293
|
CR848705.2
|
ENSDARG00000104692 |
| chr14_+_45895037 | 0.27 |
|
|
|
| chr6_-_39008833 | 0.26 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr12_+_42918686 | 0.26 |
|
|
|
| chr1_+_53353287 | 0.25 |
ENSDART00000012104
|
dla
|
deltaA |
| chr11_+_10552034 | 0.25 |
ENSDART00000157821
|
CT990625.1
|
ENSDARG00000100816 |
| chr13_+_49154065 | 0.25 |
|
|
|
| chr9_-_3524962 | 0.24 |
ENSDART00000136599
|
cybrd1
|
cytochrome b reductase 1 |
| chr6_-_40431708 | 0.24 |
ENSDART00000156005
|
si:dkey-28n18.9
|
si:dkey-28n18.9 |
| chr18_+_13589341 | 0.22 |
ENSDART00000169552
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr7_+_38740077 | 0.22 |
ENSDART00000093009
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr21_+_19409216 | 0.22 |
ENSDART00000030887
|
slc45a2
|
solute carrier family 45, member 2 |
| chr3_+_39514598 | 0.22 |
ENSDART00000156038
|
epn2
|
epsin 2 |
| chr19_+_7630847 | 0.21 |
ENSDART00000151758
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr6_-_1520414 | 0.21 |
|
|
|
| chr5_-_70948176 | 0.20 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr23_+_34392753 | 0.19 |
|
|
|
| chr19_-_44043682 | 0.17 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr16_+_31890895 | 0.16 |
ENSDART00000159821
|
atn1
|
atrophin 1 |
| chr5_-_31708973 | 0.14 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr7_+_16615154 | 0.13 |
|
|
|
| chr23_-_33434838 | 0.13 |
ENSDART00000031638
|
slc48a1a
|
solute carrier family 48 (heme transporter), member 1a |
| chr2_+_37194305 | 0.12 |
ENSDART00000176827
ENSDART00000176583 |
copa
|
coatomer protein complex, subunit alpha |
| chr25_+_25369539 | 0.11 |
ENSDART00000150537
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr21_-_10877060 | 0.11 |
|
|
|
| chr10_+_45401582 | 0.11 |
ENSDART00000159797
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr12_-_304698 | 0.11 |
|
|
|
| chr21_-_10877334 | 0.10 |
|
|
|
| chr20_+_43751193 | 0.10 |
ENSDART00000017269
|
parp1
|
poly (ADP-ribose) polymerase 1 |
| chr21_+_24948351 | 0.08 |
ENSDART00000088546
|
jam3b
|
junctional adhesion molecule 3b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0055016 | hypochord development(GO:0055016) |
| 1.0 | 4.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 1.0 | 7.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.4 | 2.4 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.4 | 2.3 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.4 | 1.4 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.4 | 2.1 | GO:2000742 | regulation of anterior head development(GO:2000742) |
| 0.3 | 1.7 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.2 | 0.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 0.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.7 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 0.7 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.2 | 1.1 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.2 | 0.5 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.2 | 1.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.2 | 1.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.6 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.2 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.1 | 2.4 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 2.2 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.1 | 1.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 1.9 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.9 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.7 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.4 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 1.4 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.1 | 2.5 | GO:2000181 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.1 | 1.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.2 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 1.8 | GO:0051693 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.0 | 0.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.7 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.3 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 1.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 2.4 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.7 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.8 | GO:0060119 | inner ear receptor cell development(GO:0060119) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.5 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 1.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.5 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 1.8 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.8 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 1.0 | GO:0009416 | response to light stimulus(GO:0009416) |
| 0.0 | 2.4 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 3.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.3 | 1.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 2.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 5.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.7 | GO:0005938 | cell cortex(GO:0005938) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.5 | 7.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 1.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 1.5 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.3 | 1.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.3 | 0.8 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 4.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 3.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 2.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 1.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 0.5 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.7 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 2.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 1.3 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 4.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 2.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 2.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 3.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 7.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 4.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.6 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.4 | 5.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 1.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |