Project
DANIO-CODE
Navigation
Downloads
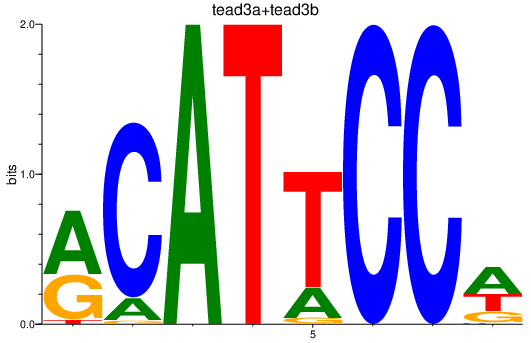
Results for tead3a+tead3b
Z-value: 6.25
Transcription factors associated with tead3a+tead3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tead3b
|
ENSDARG00000063649 | TEA domain family member 3 b |
|
tead3a
|
ENSDARG00000074321 | TEA domain family member 3 a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tead3b | dr10_dc_chr6_+_54568649_54568723 | 0.55 | 2.6e-02 | Click! |
Activity profile of tead3a+tead3b motif
Sorted Z-values of tead3a+tead3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tead3a+tead3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_29148810 | 22.40 |
ENSDART00000140350
|
thbs1b
|
thrombospondin 1b |
| chr2_-_10555152 | 21.77 |
ENSDART00000150166
|
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr6_+_15141723 | 20.74 |
ENSDART00000128090
|
ecrg4b
|
esophageal cancer related gene 4b |
| chr1_+_53286155 | 17.03 |
ENSDART00000139625
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr3_-_16387246 | 16.46 |
ENSDART00000133907
|
eps8l1
|
eps8-like1 |
| chr9_+_21466934 | 14.50 |
ENSDART00000139620
|
lats2
|
large tumor suppressor kinase 2 |
| chr5_-_70948223 | 14.35 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr6_-_39315024 | 14.29 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr13_+_22545318 | 14.13 |
ENSDART00000143312
|
zgc:193505
|
zgc:193505 |
| chr13_-_46292762 | 14.10 |
ENSDART00000149125
|
fgfr2
|
fibroblast growth factor receptor 2 |
| chr5_-_31708933 | 13.90 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr8_+_31426328 | 13.89 |
ENSDART00000135101
|
sepp1a
|
selenoprotein P, plasma, 1a |
| chr18_+_6551983 | 13.68 |
ENSDART00000160382
ENSDART00000171495 ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr2_+_36634309 | 13.61 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr15_-_20088439 | 13.33 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr23_+_10552781 | 13.27 |
|
|
|
| chr12_-_10372055 | 13.15 |
ENSDART00000052001
|
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr14_-_30363703 | 13.02 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin-like extracellular matrix protein 2a |
| chr6_+_17959219 | 12.89 |
ENSDART00000026448
|
evpla
|
envoplakin a |
| chr1_+_53353690 | 12.74 |
ENSDART00000126339
|
dla
|
deltaA |
| chr23_-_21520413 | 12.74 |
ENSDART00000044080
ENSDART00000112929 |
her12
|
hairy-related 12 |
| chr14_+_45895103 | 12.45 |
|
|
|
| chr2_-_29501573 | 12.43 |
ENSDART00000138073
|
cahz
|
carbonic anhydrase |
| chr16_-_31959111 | 12.43 |
ENSDART00000135669
|
rbfox1l
|
RNA binding protein, fox-1 homolog (C. elegans) 1-like |
| chr20_+_25441689 | 11.67 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr9_-_34146631 | 11.60 |
ENSDART00000177897
|
AL590134.2
|
ENSDARG00000107869 |
| chr3_+_36146072 | 11.55 |
ENSDART00000148444
|
zgc:86896
|
zgc:86896 |
| chr15_-_29454583 | 11.33 |
ENSDART00000145976
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr3_-_55956030 | 11.32 |
ENSDART00000158163
|
srl
|
sarcalumenin |
| chr20_+_4567653 | 11.26 |
|
|
|
| chr6_+_56157608 | 11.04 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr19_-_43037791 | 10.73 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr13_-_28543929 | 10.59 |
ENSDART00000122754
ENSDART00000057574 |
nt5c2a
|
5'-nucleotidase, cytosolic IIa |
| chr22_-_15567180 | 10.58 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr1_-_18709814 | 10.50 |
ENSDART00000145224
|
rbm47
|
RNA binding motif protein 47 |
| chr25_+_21732255 | 10.38 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr23_+_35984675 | 10.36 |
ENSDART00000053295
|
hoxc10a
|
homeobox C10a |
| chr1_-_34921848 | 9.85 |
ENSDART00000142154
|
frem3
|
Fras1 related extracellular matrix 3 |
| chr19_-_30816780 | 9.83 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr18_-_605558 | 9.80 |
ENSDART00000157564
|
wtip
|
WT1 interacting protein |
| chr1_-_19152361 | 9.79 |
ENSDART00000132958
|
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr9_+_30483319 | 9.69 |
ENSDART00000148148
|
srpx
|
sushi-repeat containing protein, X-linked |
| chr23_+_25428372 | 9.62 |
ENSDART00000147440
|
fmnl3
|
formin-like 3 |
| chr14_-_30467807 | 9.62 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr11_-_11591394 | 9.53 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr16_+_7745309 | 9.48 |
ENSDART00000081418
ENSDART00000149404 |
bves
|
blood vessel epicardial substance |
| chr11_-_42096921 | 9.45 |
|
|
|
| chr23_-_22204223 | 9.29 |
ENSDART00000079212
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr16_-_55320569 | 9.23 |
ENSDART00000156368
|
ENSDARG00000069583
|
ENSDARG00000069583 |
| chr9_+_31471391 | 9.12 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr6_+_7092350 | 9.04 |
ENSDART00000049695
ENSDART00000136088 ENSDART00000083424 ENSDART00000125912 |
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr5_+_17120453 | 9.04 |
|
|
|
| chr22_-_17652938 | 9.02 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr16_+_28819826 | 8.76 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr5_-_43334777 | 8.66 |
ENSDART00000142271
|
ENSDARG00000053091
|
ENSDARG00000053091 |
| chr19_-_11096996 | 8.65 |
ENSDART00000010997
|
tpm3
|
tropomyosin 3 |
| chr19_-_5851328 | 8.50 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr18_-_44135616 | 8.41 |
ENSDART00000087339
|
cdon
|
cell adhesion associated, oncogene regulated |
| chr12_+_27370834 | 8.40 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr1_+_46537108 | 8.33 |
|
|
|
| chr5_+_10789834 | 8.32 |
|
|
|
| chr23_-_24468191 | 8.31 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr5_-_64149931 | 8.30 |
ENSDART00000144816
|
lix1
|
limb and CNS expressed 1 |
| chr13_-_46292975 | 8.21 |
ENSDART00000167402
ENSDART00000080914 ENSDART00000080916 |
fgfr2
|
fibroblast growth factor receptor 2 |
| chr20_+_1887099 | 8.18 |
|
|
|
| chr7_-_3900223 | 8.18 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr25_-_26292712 | 8.15 |
ENSDART00000067114
|
fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr20_-_8122838 | 8.14 |
ENSDART00000113993
|
si:ch211-232i5.1
|
si:ch211-232i5.1 |
| chr16_+_46526268 | 8.12 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr20_-_49880744 | 8.06 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr4_-_24310846 | 8.05 |
ENSDART00000017443
|
celf2
|
cugbp, Elav-like family member 2 |
| chr7_+_25466534 | 7.99 |
|
|
|
| chr12_+_31558667 | 7.95 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr2_-_32372801 | 7.94 |
ENSDART00000143348
|
BX663499.1
|
ENSDARG00000093778 |
| chr19_-_33123724 | 7.91 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr17_+_1058137 | 7.85 |
|
|
|
| chr14_+_41805073 | 7.84 |
ENSDART00000074362
|
pcdh18b
|
protocadherin 18b |
| chr6_+_29420644 | 7.79 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr6_+_7309023 | 7.71 |
ENSDART00000160128
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr7_+_5372830 | 7.61 |
ENSDART00000135271
|
MYADM
|
myeloid associated differentiation marker |
| chr6_+_39373026 | 7.59 |
ENSDART00000157165
|
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr11_+_10925475 | 7.51 |
ENSDART00000064860
|
rbms1a
|
RNA binding motif, single stranded interacting protein 1a |
| chr7_-_71566426 | 7.50 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr6_+_30441419 | 7.48 |
|
|
|
| chr11_+_7148830 | 7.45 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr7_-_37587227 | 7.43 |
ENSDART00000173552
|
adcy7
|
adenylate cyclase 7 |
| chr15_+_5935952 | 7.36 |
ENSDART00000152520
ENSDART00000145827 |
sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr8_-_38126693 | 7.29 |
ENSDART00000112331
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr7_-_23878485 | 7.22 |
ENSDART00000005884
|
mmp14a
|
matrix metallopeptidase 14a (membrane-inserted) |
| chr20_-_26521700 | 7.18 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr6_+_57744081 | 7.17 |
ENSDART00000169175
|
SNTA1
|
syntrophin alpha 1 |
| chr16_+_21109486 | 7.14 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr3_-_16387615 | 7.14 |
ENSDART00000080749
ENSDART00000133824 |
eps8l1
|
eps8-like1 |
| chr17_+_32619043 | 7.13 |
ENSDART00000087565
|
eva1a
|
eva-1 homolog A (C. elegans) |
| chr16_+_39396358 | 7.11 |
|
|
|
| chr17_-_52572701 | 7.10 |
ENSDART00000060764
|
spred1
|
sprouty-related, EVH1 domain containing 1 |
| chr14_-_25301744 | 7.09 |
ENSDART00000131886
|
slc25a48
|
solute carrier family 25, member 48 |
| chr6_+_48138737 | 7.08 |
|
|
|
| chr23_+_36010592 | 7.07 |
ENSDART00000137507
|
hoxc3a
|
homeo box C3a |
| chr20_+_26639029 | 7.04 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr19_-_2079482 | 6.99 |
ENSDART00000128639
|
ENSDARG00000086326
|
ENSDARG00000086326 |
| chr19_+_5399813 | 6.97 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr5_-_18548689 | 6.96 |
ENSDART00000137802
|
fam214b
|
family with sequence similarity 214, member B |
| chr25_-_20886847 | 6.96 |
ENSDART00000110879
|
magi2b
|
membrane associated guanylate kinase, WW and PDZ domain containing 2b |
| chr23_+_20522512 | 6.92 |
ENSDART00000137294
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr24_+_7607915 | 6.85 |
ENSDART00000124409
|
cavin1b
|
caveolae associated protein 1b |
| chr1_+_32915075 | 6.76 |
ENSDART00000114384
|
znf654
|
zinc finger protein 654 |
| chr2_-_53210330 | 6.76 |
ENSDART00000024629
|
rab31
|
RAB31, member RAS oncogene family |
| chr3_-_5318206 | 6.72 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr16_+_53178713 | 6.67 |
ENSDART00000174634
|
CABZ01062546.1
|
ENSDARG00000108858 |
| chr7_-_33994315 | 6.67 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr1_+_51706536 | 6.63 |
|
|
|
| KN150131v1_+_59098 | 6.62 |
ENSDART00000175590
|
CABZ01064941.1
|
ENSDARG00000106550 |
| chr2_+_26523457 | 6.60 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr11_-_42934175 | 6.58 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr23_-_18945009 | 6.57 |
ENSDART00000080064
|
CR847953.1
|
ENSDARG00000057403 |
| chr3_-_39031305 | 6.54 |
ENSDART00000022393
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr7_+_67227810 | 6.52 |
ENSDART00000163840
|
gcshb
|
glycine cleavage system protein H (aminomethyl carrier), b |
| chr7_+_20272091 | 6.45 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr18_+_18465951 | 6.42 |
ENSDART00000165079
|
siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr5_+_32191063 | 6.30 |
ENSDART00000077189
|
ier5l
|
immediate early response 5-like |
| chr1_-_58258428 | 6.28 |
|
|
|
| chr1_-_24486146 | 6.24 |
ENSDART00000144711
|
tmem154
|
transmembrane protein 154 |
| chr13_-_33137075 | 6.20 |
ENSDART00000138096
|
oip5-as1
|
oip5 antisense RNA 1 |
| chr6_-_39315608 | 6.14 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr5_-_68270398 | 6.13 |
ENSDART00000161561
|
ENSDARG00000059093
|
ENSDARG00000059093 |
| chr23_-_7740845 | 6.13 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr23_+_28656263 | 6.10 |
ENSDART00000020296
|
nadl1.2
|
neural adhesion molecule L1.2 |
| chr16_-_29342511 | 6.08 |
ENSDART00000058870
|
rhbg
|
Rh family, B glycoprotein (gene/pseudogene) |
| KN150670v1_-_32201 | 6.05 |
|
|
|
| chr7_-_44332679 | 6.04 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr9_-_1969197 | 6.03 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr16_-_8481813 | 6.01 |
ENSDART00000051166
|
grb10b
|
growth factor receptor-bound protein 10b |
| chr24_-_16995194 | 5.97 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr1_+_16681778 | 5.96 |
|
|
|
| chr5_+_69383382 | 5.96 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr14_-_5239605 | 5.95 |
ENSDART00000054877
|
fgf24
|
fibroblast growth factor 24 |
| chr19_-_34508557 | 5.95 |
ENSDART00000160495
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr6_-_45903830 | 5.92 |
ENSDART00000025428
|
epha2a
|
eph receptor A2 a |
| chr24_-_32750010 | 5.91 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr24_+_17056023 | 5.88 |
ENSDART00000149225
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr5_+_68410884 | 5.87 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr3_-_42275696 | 5.87 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr14_-_45883839 | 5.82 |
|
|
|
| chr20_-_22478716 | 5.82 |
ENSDART00000110967
ENSDART00000011135 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr8_-_39944817 | 5.81 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr16_+_5466342 | 5.81 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr9_+_38814714 | 5.80 |
ENSDART00000114728
|
znf148
|
zinc finger protein 148 |
| chr14_+_41804998 | 5.75 |
ENSDART00000074362
|
pcdh18b
|
protocadherin 18b |
| chr14_-_46407410 | 5.72 |
ENSDART00000173078
|
si:ch211-168f7.5
|
si:ch211-168f7.5 |
| chr15_+_42440802 | 5.71 |
ENSDART00000089694
|
tiam1b
|
T-cell lymphoma invasion and metastasis 1b |
| chr13_-_46292862 | 5.71 |
ENSDART00000150061
|
fgfr2
|
fibroblast growth factor receptor 2 |
| chr7_-_41188744 | 5.71 |
ENSDART00000174087
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr15_+_42440952 | 5.70 |
ENSDART00000089694
|
tiam1b
|
T-cell lymphoma invasion and metastasis 1b |
| chr12_-_7669319 | 5.70 |
|
|
|
| chr13_-_5953330 | 5.69 |
ENSDART00000099224
|
dld
|
deltaD |
| chr4_-_12008472 | 5.67 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr1_-_8430894 | 5.60 |
ENSDART00000125596
|
si:ch211-14k19.8
|
si:ch211-14k19.8 |
| chr17_-_20938261 | 5.54 |
ENSDART00000113655
|
tmem26a
|
transmembrane protein 26a |
| chr5_+_12791779 | 5.54 |
|
|
|
| chr6_+_52064243 | 5.52 |
ENSDART00000153468
|
abraa
|
actin binding Rho activating protein a |
| chr23_+_36007936 | 5.52 |
ENSDART00000128533
|
hoxc3a
|
homeo box C3a |
| chr19_-_43038013 | 5.52 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr19_-_9603597 | 5.51 |
ENSDART00000045245
|
ing4
|
inhibitor of growth family, member 4 |
| chr1_-_37695900 | 5.49 |
ENSDART00000018533
|
hpgd
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr20_+_4567603 | 5.49 |
|
|
|
| chr1_-_19580295 | 5.49 |
ENSDART00000146084
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr24_-_16994956 | 5.49 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr14_+_34440699 | 5.48 |
ENSDART00000130469
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr24_-_6048914 | 5.48 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr17_+_38619041 | 5.43 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr9_+_26292183 | 5.35 |
|
|
|
| chr17_-_31327210 | 5.35 |
ENSDART00000154013
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr3_+_40667131 | 5.34 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr7_+_63014869 | 5.33 |
ENSDART00000161436
|
pcdh7b
|
protocadherin 7b |
| chr7_-_50244215 | 5.32 |
ENSDART00000073910
|
adamtsl5
|
ADAMTS like 5 |
| chr14_+_28136958 | 5.29 |
|
|
|
| chr17_-_26893412 | 5.22 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr7_-_26572828 | 5.22 |
ENSDART00000162241
|
si:ch211-107p11.3
|
si:ch211-107p11.3 |
| chr20_-_38546922 | 5.21 |
ENSDART00000136303
|
itpkb
|
inositol-trisphosphate 3-kinase B |
| chr2_-_32572545 | 5.16 |
ENSDART00000056641
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr3_+_23563620 | 5.15 |
ENSDART00000147022
|
hoxb7a
|
homeobox B7a |
| chr5_+_71133418 | 5.12 |
|
|
|
| chr14_-_34431528 | 5.05 |
ENSDART00000125109
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr7_-_18894639 | 5.05 |
ENSDART00000142924
|
kirrel1a
|
kin of IRRE like a |
| chr2_+_22196971 | 5.05 |
ENSDART00000140012
|
ca8
|
carbonic anhydrase VIII |
| chr7_-_23474701 | 5.03 |
ENSDART00000048050
|
ITGB1BP2
|
integrin subunit beta 1 binding protein 2 |
| chr11_+_10557530 | 5.03 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr12_-_26339002 | 5.01 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr21_+_26660833 | 5.00 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr21_+_13063614 | 5.00 |
|
|
|
| chr19_+_46380082 | 5.00 |
ENSDART00000158781
|
si:ch211-153f2.7
|
si:ch211-153f2.7 |
| chr6_-_43806125 | 4.97 |
|
|
|
| chr25_-_23428527 | 4.95 |
ENSDART00000062930
|
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr5_-_31301280 | 4.94 |
ENSDART00000141446
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr15_-_26911132 | 4.94 |
ENSDART00000077582
|
pitpnm3
|
PITPNM family member 3 |
| chr4_+_11312432 | 4.94 |
ENSDART00000051792
|
sema3aa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Aa |
| chr17_-_39237265 | 4.90 |
ENSDART00000050534
|
crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr9_+_11311198 | 4.90 |
ENSDART00000110691
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr7_+_19858190 | 4.88 |
ENSDART00000179395
ENSDART00000113793 ENSDART00000169603 |
zgc:114045
|
zgc:114045 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.3 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 4.6 | 37.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 4.3 | 17.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 3.3 | 13.3 | GO:0055016 | hypochord development(GO:0055016) |
| 3.1 | 18.5 | GO:0030104 | water homeostasis(GO:0030104) |
| 2.7 | 18.8 | GO:0072132 | mesenchyme morphogenesis(GO:0072132) |
| 2.6 | 13.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 2.4 | 7.1 | GO:2001017 | regulation of retrograde axon cargo transport(GO:2001017) |
| 2.0 | 6.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 2.0 | 14.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 1.9 | 7.7 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 1.8 | 7.3 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 1.8 | 14.5 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 1.8 | 7.1 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 1.7 | 10.5 | GO:2000742 | regulation of anterior head development(GO:2000742) |
| 1.6 | 9.9 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 1.6 | 9.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 1.6 | 9.6 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 1.6 | 4.7 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 1.5 | 5.9 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 1.4 | 5.7 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 1.4 | 9.8 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 1.4 | 15.1 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 1.3 | 4.0 | GO:0061011 | hepatic duct development(GO:0061011) |
| 1.3 | 20.8 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 1.2 | 6.2 | GO:0071632 | optomotor response(GO:0071632) |
| 1.2 | 4.8 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 1.2 | 5.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.2 | 4.6 | GO:0010481 | ventriculo bulbo valve development(GO:0003173) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 1.1 | 4.5 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.1 | 4.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 1.1 | 6.5 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 1.0 | 7.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 1.0 | 8.2 | GO:0086003 | cardiac muscle cell contraction(GO:0086003) |
| 1.0 | 6.0 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 1.0 | 4.9 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 1.0 | 5.8 | GO:0014831 | gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.9 | 11.0 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.9 | 17.1 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.9 | 11.4 | GO:0030210 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.9 | 3.5 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.9 | 5.2 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.9 | 3.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.8 | 7.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.8 | 9.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) regulation of protein deacetylation(GO:0090311) |
| 0.8 | 4.1 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic G1 DNA damage checkpoint(GO:0031571) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.8 | 2.5 | GO:0050854 | regulation of antigen receptor-mediated signaling pathway(GO:0050854) regulation of B cell receptor signaling pathway(GO:0050855) positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.8 | 4.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.8 | 3.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.8 | 27.5 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.7 | 3.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.7 | 6.0 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.7 | 7.4 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.7 | 7.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.7 | 5.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.7 | 3.5 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.7 | 2.8 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.7 | 3.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.7 | 2.7 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.7 | 4.7 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.6 | 9.0 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.6 | 1.9 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.6 | 22.4 | GO:2000181 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.6 | 2.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.6 | 2.4 | GO:0003315 | heart rudiment formation(GO:0003315) cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.6 | 6.0 | GO:0061458 | gonad development(GO:0008406) negative regulation of endodermal cell fate specification(GO:0042664) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.6 | 4.0 | GO:0019835 | cytolysis(GO:0019835) |
| 0.6 | 11.7 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.5 | 11.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.5 | 1.6 | GO:0030431 | sleep(GO:0030431) endocrine process(GO:0050886) |
| 0.5 | 8.9 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.5 | 1.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.5 | 6.0 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.5 | 2.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.5 | 7.3 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.5 | 4.4 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.5 | 8.4 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.5 | 1.8 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.4 | 3.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.4 | 2.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.4 | 10.4 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.4 | 5.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.4 | 6.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.4 | 2.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.4 | 10.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.4 | 15.5 | GO:0030835 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.4 | 9.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.4 | 3.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.4 | 3.9 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.4 | 3.9 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.4 | 1.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.4 | 4.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.4 | 2.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.4 | 7.0 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.4 | 1.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 8.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.3 | 9.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.3 | 2.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.3 | 4.9 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.3 | 1.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 5.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.3 | 8.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.3 | 10.8 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.3 | 4.1 | GO:0009746 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.3 | 7.0 | GO:0003146 | heart jogging(GO:0003146) |
| 0.3 | 4.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.3 | 3.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 5.1 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.3 | 8.5 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.2 | 1.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 1.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 7.0 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.2 | 2.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 27.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 15.9 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.2 | 15.4 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.2 | 5.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 | 2.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 1.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 0.8 | GO:0090036 | positive regulation of protein kinase A signaling(GO:0010739) bleb assembly(GO:0032060) regulation of protein kinase C signaling(GO:0090036) slow muscle cell migration(GO:1904969) |
| 0.2 | 2.1 | GO:0030168 | platelet activation(GO:0030168) |
| 0.2 | 2.7 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.2 | 10.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 11.0 | GO:0051402 | neuron apoptotic process(GO:0051402) |
| 0.2 | 3.1 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.2 | 0.7 | GO:0070073 | regulation of melanocyte differentiation(GO:0045634) clustering of voltage-gated calcium channels(GO:0070073) |
| 0.2 | 27.8 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.2 | 1.7 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.2 | 2.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 0.5 | GO:0060911 | cardiac cell fate commitment(GO:0060911) |
| 0.2 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 0.9 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.2 | 14.4 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.1 | 5.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 4.1 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.1 | 4.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 11.8 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.1 | 0.6 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 7.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 5.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 7.7 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.1 | 4.1 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 1.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 18.5 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.1 | 5.9 | GO:0021782 | glial cell development(GO:0021782) |
| 0.1 | 1.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 2.8 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.1 | 4.4 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 1.7 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 0.5 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.5 | GO:0032273 | positive regulation of protein polymerization(GO:0032273) |
| 0.1 | 13.1 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
| 0.1 | 1.1 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 4.9 | GO:0045165 | cell fate commitment(GO:0045165) |
| 0.1 | 3.3 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 3.5 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.1 | 6.4 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 1.5 | GO:0032956 | regulation of actin cytoskeleton organization(GO:0032956) |
| 0.0 | 2.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.1 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 2.0 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 7.9 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.8 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.9 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.1 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 1.4 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 2.3 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 1.7 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 1.2 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 3.4 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 3.1 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.3 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 15.5 | GO:0008091 | spectrin(GO:0008091) |
| 2.4 | 26.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.4 | 18.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.2 | 4.8 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 1.2 | 9.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 1.2 | 25.5 | GO:0045095 | keratin filament(GO:0045095) |
| 1.1 | 8.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 1.1 | 20.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.9 | 15.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.8 | 7.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.7 | 7.5 | GO:0031430 | M band(GO:0031430) |
| 0.6 | 7.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.6 | 1.8 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.5 | 5.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.5 | 1.8 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 2.8 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.4 | 5.1 | GO:0045178 | basal part of cell(GO:0045178) basal cortex(GO:0045180) |
| 0.3 | 19.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 17.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.3 | 6.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 20.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.3 | 16.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 9.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.3 | 20.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.3 | 2.3 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.3 | 60.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 5.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 3.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 2.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 14.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 6.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 6.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 4.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.2 | 2.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 14.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 1.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 9.0 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 12.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 8.0 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 68.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 65.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 4.7 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 7.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 2.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 8.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 3.8 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 6.5 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.1 | 5.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 5.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 3.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 5.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 15.4 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 10.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 9.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 9.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 1.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 7.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 16.9 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.8 | GO:0008465 | glycerate dehydrogenase activity(GO:0008465) |
| 2.6 | 12.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 2.4 | 9.8 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 2.3 | 7.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 2.3 | 7.0 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 2.2 | 26.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.9 | 7.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 1.9 | 5.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 1.8 | 28.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.4 | 27.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 1.3 | 10.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 1.2 | 6.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.2 | 4.6 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.1 | 11.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 1.1 | 12.6 | GO:0005518 | collagen binding(GO:0005518) |
| 1.1 | 18.9 | GO:0005112 | Notch binding(GO:0005112) |
| 1.0 | 2.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 1.0 | 5.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.0 | 14.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 1.0 | 5.0 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 1.0 | 9.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.9 | 3.6 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.9 | 7.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.9 | 7.1 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.9 | 2.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.9 | 6.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.9 | 9.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.9 | 8.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.8 | 2.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.8 | 4.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.8 | 3.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.8 | 22.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.7 | 3.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.7 | 16.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.7 | 2.8 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.7 | 9.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.7 | 7.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.7 | 4.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.6 | 10.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.6 | 1.8 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.6 | 5.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.6 | 8.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.5 | 6.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.5 | 1.6 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.5 | 5.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.5 | 4.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.5 | 4.8 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.5 | 1.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 1.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.4 | 2.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.4 | 5.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.4 | 2.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.4 | 5.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.4 | 15.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 2.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.4 | 1.9 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.4 | 16.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.4 | 4.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 2.5 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 3.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 5.8 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 2.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 1.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 70.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.3 | 3.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 13.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.3 | 4.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 3.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 4.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 3.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 7.4 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.2 | 3.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 5.0 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 6.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 22.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.2 | 1.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 3.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.2 | 14.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 15.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.9 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.1 | 3.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 4.4 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 2.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 3.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 24.4 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 4.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 2.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 24.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 4.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 15.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 5.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 7.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 15.5 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 1.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 15.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 9.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 18.6 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.1 | 4.7 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.1 | 5.7 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.1 | 1.0 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 2.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 6.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 6.4 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.1 | 67.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 2.4 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 2.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 4.0 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 29.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 7.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 2.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 9.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 5.5 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 2.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 7.9 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 3.5 | GO:0038023 | signaling receptor activity(GO:0038023) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.8 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 27.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 2.5 | 7.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.7 | 5.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.7 | 5.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.7 | 10.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.5 | 28.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.5 | 14.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.4 | 15.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.4 | 18.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.4 | 4.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.4 | 4.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.3 | 7.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.3 | 11.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 4.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 10.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 2.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 3.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.2 | 5.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 4.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 6.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 8.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 12.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 8.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 28.0 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 2.2 | 15.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 2.0 | 16.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 1.9 | 7.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 1.2 | 8.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 1.0 | 15.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.9 | 5.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.9 | 14.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.8 | 16.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.8 | 2.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.7 | 5.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.7 | 5.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.6 | 5.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.6 | 5.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 6.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.5 | 5.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.4 | 1.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.4 | 12.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.4 | 11.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 2.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 3.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 6.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 2.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 2.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 5.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.8 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 4.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 3.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.8 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 2.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 4.7 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |