Project
DANIO-CODE
Navigation
Downloads
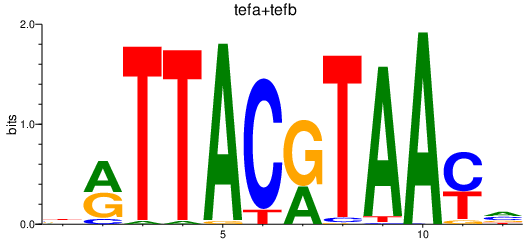
Results for tefa+tefb
Z-value: 0.56
Transcription factors associated with tefa+tefb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tefa
|
ENSDARG00000039117 | TEF transcription factor, PAR bZIP family member a |
|
tefb
|
ENSDARG00000098103 | TEF transcription factor, PAR bZIP family member b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tefa | dr10_dc_chr12_-_19030156_19030162 | -0.85 | 2.7e-05 | Click! |
| tefb | dr10_dc_chr3_-_5157662_5157883 | -0.62 | 1.1e-02 | Click! |
Activity profile of tefa+tefb motif
Sorted Z-values of tefa+tefb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tefa+tefb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_45103102 | 2.07 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr3_-_31672763 | 1.73 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr22_-_13139894 | 1.58 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr16_+_46145286 | 1.34 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr4_-_12726432 | 1.34 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr4_-_5294280 | 1.28 |
ENSDART00000178921
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr17_-_52020689 | 1.28 |
ENSDART00000019766
|
tgfb3
|
transforming growth factor, beta 3 |
| chr20_-_44598129 | 1.22 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr19_+_5073991 | 1.21 |
ENSDART00000151050
|
ENSDARG00000011076
|
ENSDARG00000011076 |
| chr23_+_22977031 | 1.02 |
|
|
|
| chr3_-_21217799 | 1.00 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr15_-_40391882 | 0.98 |
|
|
|
| chr25_-_18234069 | 0.97 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr3_+_29583029 | 0.96 |
|
|
|
| chr19_-_46058954 | 0.94 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr23_+_4083697 | 0.94 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr20_-_47521258 | 0.92 |
|
|
|
| chr5_-_21497169 | 0.90 |
ENSDART00000143878
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr16_+_17705704 | 0.88 |
|
|
|
| chr21_+_21337628 | 0.87 |
ENSDART00000136325
|
rtn2b
|
reticulon 2b |
| chr8_+_8673307 | 0.86 |
ENSDART00000110854
|
elk1
|
ELK1, member of ETS oncogene family |
| chr24_-_39884167 | 0.85 |
ENSDART00000087441
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr3_-_11892052 | 0.85 |
|
|
|
| chr8_+_42992323 | 0.82 |
ENSDART00000048819
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr2_+_24521514 | 0.81 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr5_+_68943914 | 0.80 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr23_+_4083748 | 0.79 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr11_-_23206021 | 0.79 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr19_-_46058895 | 0.77 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr24_+_18804086 | 0.77 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr8_+_50498398 | 0.76 |
|
|
|
| chr20_+_38298893 | 0.64 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr19_-_22914516 | 0.63 |
|
|
|
| chr22_-_11408859 | 0.63 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr14_+_30568242 | 0.62 |
|
|
|
| chr13_+_24703802 | 0.60 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr9_+_33979089 | 0.60 |
ENSDART00000144623
|
kdm6a
|
lysine (K)-specific demethylase 6A |
| chr15_-_21003820 | 0.60 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr5_-_38224191 | 0.59 |
ENSDART00000147121
|
cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr6_+_16341787 | 0.57 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
| chr4_-_5293599 | 0.52 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr4_-_12915292 | 0.50 |
ENSDART00000067135
|
msrb3
|
methionine sulfoxide reductase B3 |
| chr2_+_289138 | 0.50 |
ENSDART00000157246
|
znf1008
|
zinc finger protein 1008 |
| chr17_-_43475725 | 0.49 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr17_-_16079935 | 0.49 |
|
|
|
| chr10_+_18953671 | 0.47 |
ENSDART00000135786
|
bnip3lb
|
BCL2/adenovirus E1B interacting protein 3-like b |
| chr17_-_8111181 | 0.47 |
ENSDART00000149873
ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr20_-_45757288 | 0.45 |
ENSDART00000124582
ENSDART00000100290 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr5_-_20527105 | 0.45 |
ENSDART00000133461
|
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr20_-_54942216 | 0.44 |
|
|
|
| chr5_+_22204129 | 0.44 |
|
|
|
| KN150708v1_-_31573 | 0.43 |
ENSDART00000160040
|
slc16a6a
|
solute carrier family 16, member 6a |
| chr20_+_38298755 | 0.42 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr7_+_10346939 | 0.42 |
|
|
|
| chr14_+_1152700 | 0.42 |
ENSDART00000125203
|
hopx
|
HOP homeobox |
| chr3_-_28534446 | 0.40 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr21_+_25588388 | 0.39 |
ENSDART00000134678
|
ENSDARG00000078256
|
ENSDARG00000078256 |
| chr5_+_14850708 | 0.37 |
ENSDART00000140990
ENSDART00000137287 |
pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr8_-_37217197 | 0.37 |
ENSDART00000178556
|
rbm39b
|
RNA binding motif protein 39b |
| chr8_-_29842345 | 0.36 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr17_-_17744885 | 0.36 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr25_-_211879 | 0.34 |
ENSDART00000021614
|
srpk2
|
SRSF protein kinase 2 |
| chr22_-_18466781 | 0.33 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr10_-_25629613 | 0.33 |
ENSDART00000131640
|
tiam1a
|
T-cell lymphoma invasion and metastasis 1a |
| chr15_+_28370102 | 0.32 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr23_+_34037058 | 0.31 |
ENSDART00000140666
|
prosc
|
pyridoxal phosphate binding protein |
| chr12_-_26291790 | 0.31 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr25_-_10992022 | 0.29 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr21_+_4345150 | 0.27 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr16_+_17706003 | 0.27 |
|
|
|
| chr17_-_20959288 | 0.25 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr25_+_15257730 | 0.24 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr17_-_31290969 | 0.22 |
|
|
|
| chr19_-_46058123 | 0.21 |
|
|
|
| chr20_+_23184685 | 0.20 |
ENSDART00000132093
|
usp46
|
ubiquitin specific peptidase 46 |
| chr19_+_8693914 | 0.18 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr3_+_24564249 | 0.18 |
ENSDART00000111769
|
mkl1a
|
megakaryoblastic leukemia (translocation) 1a |
| chr12_+_18596599 | 0.17 |
ENSDART00000152935
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr12_-_31369772 | 0.17 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr14_+_4044357 | 0.16 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr18_-_2580718 | 0.13 |
|
|
|
| chr5_+_18837448 | 0.13 |
|
|
|
| chr10_+_32949485 | 0.11 |
|
|
|
| chr24_+_28481885 | 0.09 |
ENSDART00000079770
ENSDART00000147063 |
abca4a
|
ATP-binding cassette, sub-family A (ABC1), member 4a |
| chr18_-_46831863 | 0.07 |
|
|
|
| chr19_+_7630847 | 0.06 |
ENSDART00000151758
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr8_+_21192955 | 0.06 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr6_+_16342020 | 0.00 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0022602 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.3 | 1.0 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 0.2 | 1.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.4 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.1 | 0.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.8 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 1.3 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.6 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.4 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.3 | GO:0005902 | microvillus(GO:0005902) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.2 | 0.8 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 1.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.5 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 1.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 2.1 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.1 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 0.9 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |