Project
DANIO-CODE
Navigation
Downloads
Results for tfap2a+tfap2c
Z-value: 0.29
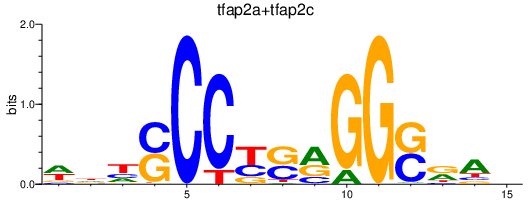
Transcription factors associated with tfap2a+tfap2c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2c
|
ENSDARG00000040606 | transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
|
tfap2a
|
ENSDARG00000059279 | transcription factor AP-2 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfap2a | dr10_dc_chr24_-_8589388_8589393 | 0.46 | 7.5e-02 | Click! |
| tfap2c | dr10_dc_chr6_+_56163589_56163640 | 0.35 | 1.8e-01 | Click! |
Activity profile of tfap2a+tfap2c motif
Sorted Z-values of tfap2a+tfap2c motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2a+tfap2c
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_17451381 | 0.48 |
ENSDART00000167602
|
spaca4l
|
sperm acrosome associated 4 like |
| chr11_-_5793 | 0.45 |
|
|
|
| chr23_-_27226280 | 0.35 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr7_-_33994315 | 0.31 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr23_-_27226387 | 0.27 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr10_-_5135388 | 0.26 |
ENSDART00000108587
|
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr19_-_5452918 | 0.26 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr14_-_27703812 | 0.25 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr5_-_62116975 | 0.24 |
ENSDART00000144469
|
adora2b
|
adenosine A2b receptor |
| chr23_-_27721250 | 0.23 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr13_-_28543929 | 0.21 |
ENSDART00000122754
ENSDART00000057574 |
nt5c2a
|
5'-nucleotidase, cytosolic IIa |
| chr18_+_34205362 | 0.18 |
ENSDART00000130831
|
gmps
|
guanine monophosphate synthase |
| chr5_-_37851313 | 0.18 |
ENSDART00000136428
ENSDART00000112487 |
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr6_-_19928053 | 0.18 |
ENSDART00000161257
|
plxnb1b
|
plexin b1b |
| chr16_-_7876036 | 0.18 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr23_-_27721202 | 0.17 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr25_+_35855292 | 0.17 |
ENSDART00000152801
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr8_+_39525254 | 0.14 |
|
|
|
| chr1_+_45782078 | 0.14 |
ENSDART00000166028
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr3_+_26113393 | 0.13 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr16_-_5243402 | 0.13 |
ENSDART00000131876
|
ttk
|
ttk protein kinase |
| chr14_-_46476712 | 0.13 |
ENSDART00000173209
ENSDART00000164837 |
ehd1a
|
EH-domain containing 1a |
| chr14_-_46477165 | 0.13 |
ENSDART00000173209
ENSDART00000164837 |
ehd1a
|
EH-domain containing 1a |
| chr23_-_21288972 | 0.12 |
ENSDART00000143206
|
megf6a
|
multiple EGF-like-domains 6a |
| chr21_+_3556841 | 0.12 |
|
|
|
| chr3_-_55073857 | 0.12 |
ENSDART00000082944
|
dock6
|
dedicator of cytokinesis 6 |
| chr14_+_14501263 | 0.11 |
ENSDART00000169235
|
fhdc2
|
FH2 domain containing 2 |
| chr21_-_39013447 | 0.11 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr3_+_26113651 | 0.11 |
ENSDART00000103734
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr24_+_31437128 | 0.11 |
ENSDART00000172335
|
cpne3
|
copine III |
| chr1_+_15855776 | 0.11 |
ENSDART00000048855
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr20_-_3873641 | 0.11 |
ENSDART00000113132
|
gnpat
|
glyceronephosphate O-acyltransferase |
| chr19_-_5453469 | 0.11 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr7_+_32422659 | 0.11 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr17_+_25503946 | 0.11 |
|
|
|
| chr20_-_20412830 | 0.10 |
ENSDART00000114779
|
ENSDARG00000079369
|
ENSDARG00000079369 |
| chr25_-_5039781 | 0.09 |
|
|
|
| chr11_+_37639045 | 0.08 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr11_+_31061296 | 0.08 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr20_-_2708593 | 0.08 |
ENSDART00000145730
|
akirin2
|
akirin 2 |
| chr4_-_21745163 | 0.08 |
ENSDART00000139962
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr5_+_61136704 | 0.07 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr25_-_4376129 | 0.07 |
|
|
|
| chr19_+_24741199 | 0.07 |
ENSDART00000090081
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr14_-_29498646 | 0.07 |
|
|
|
| chr16_+_24697776 | 0.06 |
|
|
|
| chr17_+_8765158 | 0.06 |
|
|
|
| chr10_-_210681 | 0.06 |
ENSDART00000059476
|
psmg1
|
proteasome (prosome, macropain) assembly chaperone 1 |
| chr10_+_5135537 | 0.06 |
ENSDART00000132627
ENSDART00000082295 |
zgc:113274
|
zgc:113274 |
| chr3_+_34716635 | 0.06 |
ENSDART00000055263
|
itga3a
|
integrin, alpha 3a |
| chr1_+_25773300 | 0.05 |
ENSDART00000011809
|
uso1
|
USO1 vesicle transport factor |
| chr14_-_46476881 | 0.05 |
ENSDART00000173209
ENSDART00000164837 |
ehd1a
|
EH-domain containing 1a |
| chr14_-_549816 | 0.04 |
ENSDART00000168951
|
fgf2
|
fibroblast growth factor 2 |
| chr8_+_48953831 | 0.04 |
ENSDART00000132035
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr3_-_56969897 | 0.03 |
|
|
|
| chr19_-_30923513 | 0.03 |
ENSDART00000088760
|
bag6l
|
BCL2-associated athanogene 6, like |
| chr16_-_24697750 | 0.03 |
ENSDART00000163305
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr10_+_44847446 | 0.03 |
|
|
|
| chr7_-_15004167 | 0.02 |
ENSDART00000030009
|
ENSDARG00000023868
|
ENSDARG00000023868 |
| chr3_-_40407003 | 0.02 |
|
|
|
| chr8_+_48953644 | 0.02 |
ENSDART00000132035
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr20_-_2708686 | 0.02 |
ENSDART00000145730
|
akirin2
|
akirin 2 |
| chr20_-_2708839 | 0.02 |
ENSDART00000152120
|
akirin2
|
akirin 2 |
| chr3_-_26675055 | 0.02 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr20_-_15181042 | 0.01 |
ENSDART00000175585
|
prrc2c
|
proline-rich coiled-coil 2C |
| chr18_-_15582896 | 0.01 |
ENSDART00000172690
ENSDART00000159915 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr19_+_24741351 | 0.01 |
ENSDART00000090081
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr8_-_10966975 | 0.01 |
ENSDART00000020116
|
trim33
|
tripartite motif containing 33 |
| chr5_-_37850885 | 0.01 |
ENSDART00000136428
ENSDART00000112487 |
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr10_+_40330533 | 0.01 |
|
|
|
| chr4_-_16356071 | 0.00 |
ENSDART00000079521
|
kera
|
keratocan |
| chr3_+_34716733 | 0.00 |
ENSDART00000055263
|
itga3a
|
integrin, alpha 3a |
| chr20_-_34389652 | 0.00 |
ENSDART00000144705
ENSDART00000020389 |
hmcn1
|
hemicentin 1 |
| chr2_-_22173254 | 0.00 |
ENSDART00000137538
|
rab2a
|
RAB2A, member RAS oncogene family |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.2 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.2 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.0 | 0.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |