Project
DANIO-CODE
Navigation
Downloads
Results for tfap2b
Z-value: 0.37
Transcription factors associated with tfap2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2b
|
ENSDARG00000012667 | transcription factor AP-2 beta |
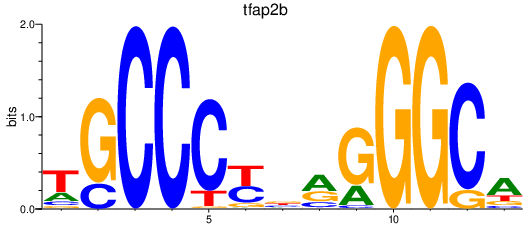
Activity profile of tfap2b motif
Sorted Z-values of tfap2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_92920 | 0.77 |
ENSDART00000099074
|
si:zfos-411a11.2
|
si:zfos-411a11.2 |
| chr15_-_104331 | 0.72 |
ENSDART00000164323
ENSDART00000161218 |
cyp2y3
|
cytochrome P450, family 2, subfamily Y, polypeptide 3 |
| chr11_-_18818010 | 0.51 |
|
|
|
| chr1_-_48900185 | 0.48 |
ENSDART00000113072
|
zgc:163136
|
zgc:163136 |
| chr21_-_30378764 | 0.37 |
ENSDART00000136247
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr4_-_29817874 | 0.36 |
|
|
|
| chr4_-_62271066 | 0.31 |
ENSDART00000162955
|
znf1091
|
zinc finger protein 1091 |
| chr16_-_21238288 | 0.28 |
ENSDART00000139737
|
cbx3b
|
chromobox homolog 3b |
| chr11_-_9479592 | 0.26 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr11_-_9479689 | 0.24 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr10_-_5135388 | 0.24 |
ENSDART00000108587
|
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr7_+_40879017 | 0.23 |
|
|
|
| chr1_-_39177682 | 0.22 |
ENSDART00000035739
|
tmem134
|
transmembrane protein 134 |
| chr17_-_26849495 | 0.22 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr7_-_8635687 | 0.22 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr11_-_36853345 | 0.21 |
ENSDART00000172074
|
NINJ1
|
ninjurin 1 |
| chr19_-_19138287 | 0.18 |
|
|
|
| chr9_-_42894582 | 0.17 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr8_+_39525254 | 0.17 |
|
|
|
| chr12_+_30245620 | 0.17 |
ENSDART00000153116
ENSDART00000152900 |
ENSDARG00000020224
|
ENSDARG00000020224 |
| chr1_+_13386937 | 0.17 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr15_-_104637 | 0.15 |
ENSDART00000164323
ENSDART00000161218 |
cyp2y3
|
cytochrome P450, family 2, subfamily Y, polypeptide 3 |
| chr2_-_42566339 | 0.15 |
ENSDART00000121452
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr12_-_8920454 | 0.14 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr12_+_5673442 | 0.14 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr15_-_4154407 | 0.13 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr22_+_470536 | 0.13 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr4_+_5826813 | 0.12 |
ENSDART00000055414
|
pex26
|
peroxisomal biogenesis factor 26 |
| chr13_+_36459625 | 0.11 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr23_-_1008307 | 0.11 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr12_-_8920541 | 0.11 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr8_-_52728470 | 0.11 |
|
|
|
| chr9_-_14916484 | 0.10 |
|
|
|
| chr10_+_1812271 | 0.09 |
|
|
|
| chr18_+_24932972 | 0.09 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr13_+_36459562 | 0.09 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr21_-_34891991 | 0.09 |
ENSDART00000065337
|
kif20a
|
kinesin family member 20A |
| chr7_-_20201693 | 0.09 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr3_+_15122138 | 0.08 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr7_+_5853590 | 0.07 |
ENSDART00000123660
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr2_-_30340646 | 0.07 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr9_-_15453448 | 0.07 |
ENSDART00000124346
ENSDART00000010521 ENSDART00000103753 |
fn1a
|
fibronectin 1a |
| chr8_+_29953399 | 0.07 |
ENSDART00000149372
ENSDART00000007640 |
ptch1
ptch1
|
patched 1 patched 1 |
| chr21_+_28687401 | 0.06 |
ENSDART00000138017
|
puraa
|
purine-rich element binding protein Aa |
| chr17_+_52780715 | 0.05 |
|
|
|
| chr14_+_10797695 | 0.05 |
ENSDART00000161311
|
NEXMIF (1 of many)
|
neurite extension and migration factor |
| chr10_+_5135537 | 0.04 |
ENSDART00000132627
ENSDART00000082295 |
zgc:113274
|
zgc:113274 |
| chr12_+_35018663 | 0.04 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr14_+_49180737 | 0.04 |
|
|
|
| chr7_-_38299544 | 0.04 |
ENSDART00000041055
|
celf1
|
cugbp, Elav-like family member 1 |
| chr16_+_54350976 | 0.04 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr7_+_1443102 | 0.04 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr16_-_2269074 | 0.03 |
|
|
|
| chr5_+_33011105 | 0.03 |
|
|
|
| chr10_+_6381994 | 0.03 |
ENSDART00000170548
|
ENSDARG00000100633
|
ENSDARG00000100633 |
| chr23_+_39569262 | 0.02 |
ENSDART00000031616
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr7_-_57680717 | 0.02 |
|
|
|
| chr11_+_18817797 | 0.02 |
|
|
|
| chr23_-_1008468 | 0.01 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr14_+_49180887 | 0.01 |
|
|
|
| chr25_-_31952245 | 0.01 |
ENSDART00000153892
|
cep152
|
centrosomal protein 152 |
| chr14_+_49180922 | 0.00 |
|
|
|
| chr2_-_31817355 | 0.00 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr2_-_44329092 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003131 | mesodermal-endodermal cell signaling(GO:0003131) response to vitamin A(GO:0033189) cellular response to vitamin A(GO:0071299) retinoic acid receptor signaling pathway involved in somitogenesis(GO:0090242) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) actin filament debranching(GO:0071846) |
| 0.0 | 0.9 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0051117 | ATPase binding(GO:0051117) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |