Project
DANIO-CODE
Navigation
Downloads
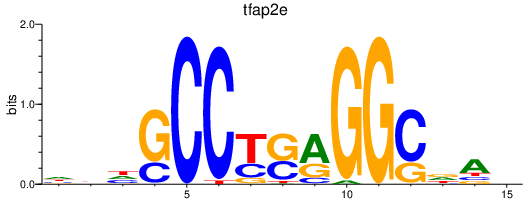
Results for tfap2e
Z-value: 0.55
Transcription factors associated with tfap2e
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2e
|
ENSDARG00000008861 | transcription factor AP-2 epsilon |
Activity profile of tfap2e motif
Sorted Z-values of tfap2e motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2e
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_24148599 | 1.70 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr22_+_12327849 | 1.19 |
ENSDART00000178678
|
FO704572.1
|
ENSDARG00000106890 |
| chr6_-_39315024 | 1.04 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr14_-_26406720 | 0.97 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr19_-_23037220 | 0.89 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr2_+_36634309 | 0.88 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr21_-_39013447 | 0.84 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr23_-_27721250 | 0.82 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr9_+_33167554 | 0.79 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| KN150230v1_-_36595 | 0.76 |
|
|
|
| chr5_-_37850885 | 0.67 |
ENSDART00000136428
ENSDART00000112487 |
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr7_-_72500748 | 0.65 |
ENSDART00000160523
|
CU929444.1
|
ENSDARG00000099109 |
| chr19_-_35513216 | 0.65 |
|
|
|
| chr8_+_54171992 | 0.64 |
ENSDART00000122692
|
CABZ01112317.1
|
ENSDARG00000086057 |
| chr5_-_37851313 | 0.60 |
ENSDART00000136428
ENSDART00000112487 |
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr19_-_9553977 | 0.57 |
ENSDART00000045565
|
vamp1
|
vesicle-associated membrane protein 1 |
| chr25_-_5039781 | 0.57 |
|
|
|
| chr15_+_21399127 | 0.51 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr23_-_27721202 | 0.48 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr19_+_17451381 | 0.45 |
ENSDART00000167602
|
spaca4l
|
sperm acrosome associated 4 like |
| chr17_+_25503946 | 0.44 |
|
|
|
| chr3_+_29380133 | 0.44 |
ENSDART00000123619
|
rac2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr6_-_39315608 | 0.43 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr19_-_5453469 | 0.41 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr15_+_21399299 | 0.40 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr16_+_36952821 | 0.40 |
ENSDART00000160645
|
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr20_-_15181042 | 0.40 |
ENSDART00000175585
|
prrc2c
|
proline-rich coiled-coil 2C |
| chr25_+_35855292 | 0.40 |
ENSDART00000152801
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr13_+_24148474 | 0.39 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr19_+_47720261 | 0.38 |
ENSDART00000136647
|
ext1c
|
exostoses (multiple) 1c |
| chr19_-_27878747 | 0.35 |
ENSDART00000137638
|
si:ch73-25f10.6
|
si:ch73-25f10.6 |
| chr10_-_5135388 | 0.33 |
ENSDART00000108587
|
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr3_-_54352640 | 0.30 |
ENSDART00000128041
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr24_+_33697332 | 0.29 |
|
|
|
| chr5_-_62116975 | 0.28 |
ENSDART00000144469
|
adora2b
|
adenosine A2b receptor |
| chr11_+_31061296 | 0.28 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr4_-_2065017 | 0.28 |
ENSDART00000087835
|
ano6
|
anoctamin 6 |
| chr23_-_5429419 | 0.27 |
ENSDART00000159105
|
CR936540.3
|
ENSDARG00000101435 |
| chr20_-_20412830 | 0.26 |
ENSDART00000114779
|
ENSDARG00000079369
|
ENSDARG00000079369 |
| chr7_-_15004167 | 0.23 |
ENSDART00000030009
|
ENSDARG00000023868
|
ENSDARG00000023868 |
| chr19_-_27878556 | 0.23 |
ENSDART00000137638
|
si:ch73-25f10.6
|
si:ch73-25f10.6 |
| chr3_-_56969897 | 0.23 |
|
|
|
| chr10_+_5135537 | 0.23 |
ENSDART00000132627
ENSDART00000082295 |
zgc:113274
|
zgc:113274 |
| chr8_-_7618945 | 0.20 |
ENSDART00000040672
|
mecp2
|
methyl CpG binding protein 2 |
| chr3_+_25859153 | 0.20 |
ENSDART00000007398
|
gcat
|
glycine C-acetyltransferase |
| chr17_+_34238628 | 0.19 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr24_-_21758244 | 0.19 |
ENSDART00000091252
|
spata13
|
spermatogenesis associated 13 |
| chr11_-_11722452 | 0.18 |
ENSDART00000161293
|
CR848705.2
|
ENSDARG00000104692 |
| chr15_+_21399228 | 0.18 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr2_+_36634251 | 0.18 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr21_+_39918070 | 0.18 |
ENSDART00000135235
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr24_+_42133471 | 0.17 |
ENSDART00000170514
|
top1mt
|
topoisomerase (DNA) I, mitochondrial |
| chr19_-_5452918 | 0.17 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr14_-_34265449 | 0.17 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr2_-_15373241 | 0.16 |
ENSDART00000163858
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr9_+_33167471 | 0.15 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr16_-_7876036 | 0.14 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr3_+_45345181 | 0.11 |
|
|
|
| chr14_-_29498646 | 0.11 |
|
|
|
| chr8_-_47156727 | 0.10 |
|
|
|
| chr7_-_2308580 | 0.08 |
ENSDART00000098148
ENSDART00000173624 |
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr17_-_15141309 | 0.07 |
ENSDART00000103405
|
gch1
|
GTP cyclohydrolase 1 |
| chr4_+_76581339 | 0.06 |
ENSDART00000174203
|
zgc:113921
|
zgc:113921 |
| chr2_-_10724196 | 0.06 |
|
|
|
| chr8_-_47156995 | 0.06 |
|
|
|
| chr4_-_21745163 | 0.06 |
ENSDART00000139962
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr13_+_24148390 | 0.06 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr20_+_22780756 | 0.04 |
|
|
|
| chr19_-_30923513 | 0.04 |
ENSDART00000088760
|
bag6l
|
BCL2-associated athanogene 6, like |
| chr2_-_22173254 | 0.04 |
ENSDART00000137538
|
rab2a
|
RAB2A, member RAS oncogene family |
| chr24_+_42133272 | 0.01 |
ENSDART00000170514
|
top1mt
|
topoisomerase (DNA) I, mitochondrial |
| chr10_+_16089472 | 0.01 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.4 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 2.1 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.3 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.3 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.8 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.1 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.6 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.2 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |