Project
DANIO-CODE
Navigation
Downloads
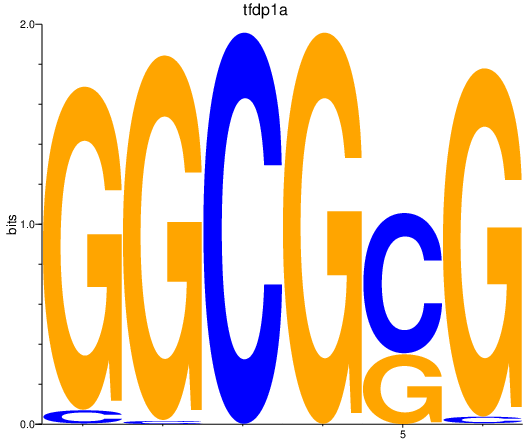
Results for tfdp1a
Z-value: 2.29
Transcription factors associated with tfdp1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfdp1a
|
ENSDARG00000019293 | transcription factor Dp-1, a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfdp1a | dr10_dc_chr9_+_35141690_35141816 | -0.25 | 3.5e-01 | Click! |
Activity profile of tfdp1a motif
Sorted Z-values of tfdp1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tfdp1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_58397256 | 3.70 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr4_+_5733160 | 3.57 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr12_-_34614931 | 3.20 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr6_+_251026 | 3.14 |
ENSDART00000042970
|
atf4a
|
activating transcription factor 4a |
| chr8_-_40287789 | 3.02 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr1_+_29054033 | 2.78 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr24_-_37680649 | 2.73 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr15_-_4322875 | 2.61 |
ENSDART00000173311
|
si:ch211-117a13.2
|
si:ch211-117a13.2 |
| chr25_+_182037 | 2.59 |
ENSDART00000156469
|
RPS17
|
ribosomal protein S17 |
| chr13_+_371940 | 2.58 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr3_-_60522527 | 2.56 |
ENSDART00000166334
|
srsf2b
|
serine/arginine-rich splicing factor 2b |
| chr15_+_29153215 | 2.56 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| KN150617v1_+_66361 | 2.52 |
ENSDART00000159503
|
fzd4
|
frizzled class receptor 4 |
| chr4_-_18606513 | 2.50 |
ENSDART00000049061
|
cdkn1ba
|
cyclin-dependent kinase inhibitor 1Ba |
| chr16_+_42993841 | 2.46 |
ENSDART00000156767
|
CR854827.4
|
ENSDARG00000097750 |
| chr12_-_47699958 | 2.45 |
ENSDART00000171932
ENSDART00000168165 ENSDART00000161985 |
HHEX
|
hematopoietically expressed homeobox |
| chr4_-_25075693 | 2.41 |
ENSDART00000025153
|
gata3
|
GATA binding protein 3 |
| chr16_-_46799406 | 2.31 |
ENSDART00000115244
|
MEX3A
|
mex-3 RNA binding family member A |
| chr2_+_25363717 | 2.30 |
ENSDART00000142601
|
stag1a
|
stromal antigen 1a |
| chr22_-_37414940 | 2.30 |
ENSDART00000104493
|
sox2
|
SRY (sex determining region Y)-box 2 |
| chr9_-_41608298 | 2.28 |
|
|
|
| chr23_-_24468191 | 2.27 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr2_+_48448974 | 2.19 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr20_+_45989418 | 2.18 |
ENSDART00000131169
|
bmp2b
|
bone morphogenetic protein 2b |
| chr4_-_2615160 | 2.17 |
ENSDART00000140760
|
e2f7
|
E2F transcription factor 7 |
| chr15_+_37039861 | 2.16 |
ENSDART00000172664
|
kirrel3l
|
kin of IRRE like 3 like |
| chr6_+_12733209 | 2.10 |
ENSDART00000167021
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr21_-_41445163 | 2.10 |
ENSDART00000170457
|
hmp19
|
HMP19 protein |
| chr7_-_6334472 | 2.03 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr4_+_1750689 | 2.02 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr3_-_55995924 | 2.01 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr5_-_66024357 | 1.98 |
|
|
|
| chr10_-_3294792 | 1.91 |
ENSDART00000109131
|
slc25a1b
|
slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b |
| chr22_+_16471319 | 1.88 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr6_+_7092350 | 1.86 |
ENSDART00000049695
ENSDART00000136088 ENSDART00000083424 ENSDART00000125912 |
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr4_+_3970874 | 1.85 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr2_-_58111727 | 1.84 |
ENSDART00000004431
ENSDART00000166845 ENSDART00000163999 |
epb41l3b
|
erythrocyte membrane protein band 4.1-like 3b |
| chr14_+_150214 | 1.81 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr6_+_56163589 | 1.79 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr2_+_42874975 | 1.78 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr10_+_25407847 | 1.77 |
ENSDART00000047541
|
bach1b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
| chr3_+_57878398 | 1.76 |
ENSDART00000047418
|
notum1a
|
notum pectinacetylesterase homolog 1a (Drosophila) |
| chr4_+_22959458 | 1.74 |
ENSDART00000036531
|
gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr20_-_158899 | 1.73 |
ENSDART00000131635
|
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr3_+_24067387 | 1.73 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr5_-_35729429 | 1.73 |
|
|
|
| chr24_+_389982 | 1.73 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr4_-_17735989 | 1.72 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr12_-_31609033 | 1.71 |
ENSDART00000153102
|
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr9_-_9693163 | 1.68 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr22_+_20436287 | 1.68 |
ENSDART00000135984
|
onecut3a
|
one cut homeobox 3a |
| chr13_+_18066369 | 1.66 |
ENSDART00000109642
|
tet1
|
tet methylcytosine dioxygenase 1 |
| chr4_+_830826 | 1.65 |
|
|
|
| chr8_+_387564 | 1.65 |
ENSDART00000167361
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr25_-_20994084 | 1.64 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| chr5_-_14148028 | 1.61 |
ENSDART00000113037
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr14_+_14759314 | 1.60 |
ENSDART00000167075
|
nkx1.2lb
|
NK1 transcription factor related 2-like,b |
| chr16_-_16433191 | 1.59 |
ENSDART00000022685
|
nradd
|
neurotrophin receptor associated death domain |
| chr5_+_6054781 | 1.58 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr21_-_34623741 | 1.57 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr23_+_24141576 | 1.57 |
|
|
|
| chr1_-_46015367 | 1.57 |
ENSDART00000074519
|
kpna3
|
karyopherin alpha 3 (importin alpha 4) |
| chr8_+_10267246 | 1.57 |
ENSDART00000159312
ENSDART00000160766 |
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr10_-_35036872 | 1.56 |
ENSDART00000170625
|
smad9
|
SMAD family member 9 |
| chr11_+_21749658 | 1.55 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr19_+_7234029 | 1.55 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr25_-_13580057 | 1.54 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr12_+_34631232 | 1.54 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr8_+_15987710 | 1.53 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr21_+_35181329 | 1.52 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr8_+_32604349 | 1.51 |
ENSDART00000126833
|
hmcn2
|
hemicentin 2 |
| chr8_-_17480730 | 1.51 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr20_-_25063937 | 1.49 |
ENSDART00000159122
|
epha7
|
eph receptor A7 |
| chr22_-_3134876 | 1.48 |
ENSDART00000159580
|
lmnb2
|
lamin B2 |
| chr23_-_12310778 | 1.48 |
ENSDART00000131256
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr16_+_6923898 | 1.48 |
ENSDART00000078306
|
arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr3_+_47826386 | 1.48 |
ENSDART00000003338
|
unkl
|
unkempt family zinc finger-like |
| chr23_-_35595271 | 1.47 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr7_-_6219627 | 1.46 |
ENSDART00000172898
|
HIST2H2AB (1 of many)
|
zgc:165551 |
| chr15_-_18425091 | 1.46 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr21_-_3301751 | 1.45 |
ENSDART00000009740
|
smad7
|
SMAD family member 7 |
| chr8_+_54171992 | 1.44 |
ENSDART00000122692
|
CABZ01112317.1
|
ENSDARG00000086057 |
| chr8_+_388003 | 1.44 |
ENSDART00000067668
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr7_-_6309265 | 1.44 |
ENSDART00000172825
|
FP325123.5
|
Histone H3.2 |
| chr18_+_5899355 | 1.42 |
|
|
|
| chr16_-_53896201 | 1.42 |
ENSDART00000179533
|
fzd1
|
frizzled class receptor 1 |
| chr15_-_20297270 | 1.41 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr4_-_824333 | 1.41 |
ENSDART00000135618
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr17_+_53224434 | 1.40 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr24_+_9335428 | 1.40 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr16_-_24697750 | 1.39 |
ENSDART00000163305
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr9_+_54692834 | 1.39 |
|
|
|
| chr5_-_24509021 | 1.39 |
ENSDART00000177338
ENSDART00000177557 ENSDART00000177478 |
emid1
|
EMI domain containing 1 |
| chr4_-_18212739 | 1.39 |
ENSDART00000169704
|
anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr11_-_11591394 | 1.38 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr5_-_40707316 | 1.37 |
ENSDART00000161932
|
npr3
|
natriuretic peptide receptor 3 |
| chr4_-_2549493 | 1.37 |
|
|
|
| chr23_-_23474703 | 1.37 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr16_-_14506846 | 1.37 |
ENSDART00000170957
|
crabp2a
|
cellular retinoic acid binding protein 2, a |
| chr25_-_22089794 | 1.35 |
ENSDART00000139110
|
pkp3a
|
plakophilin 3a |
| chr22_+_223797 | 1.32 |
|
|
|
| chr23_-_28420470 | 1.32 |
ENSDART00000145072
|
neurod4
|
neuronal differentiation 4 |
| chr2_-_30198789 | 1.32 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr24_-_35811390 | 1.31 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| KN150001v1_+_14939 | 1.31 |
|
|
|
| chr6_+_49096966 | 1.30 |
ENSDART00000141042
ENSDART00000143369 |
slc25a22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr9_+_38975508 | 1.30 |
|
|
|
| chr12_+_11042472 | 1.30 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr24_+_27469268 | 1.30 |
ENSDART00000105774
|
ek1
|
eph-like kinase 1 |
| chr1_+_23385409 | 1.30 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr10_+_4094568 | 1.29 |
ENSDART00000114533
|
mn1a
|
meningioma 1a |
| chr25_-_20283541 | 1.29 |
ENSDART00000142665
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr4_+_10065500 | 1.29 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr18_-_605558 | 1.29 |
ENSDART00000157564
|
wtip
|
WT1 interacting protein |
| chr25_-_36876940 | 1.29 |
|
|
|
| chr6_+_56157608 | 1.29 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr9_-_44493074 | 1.27 |
ENSDART00000167685
|
neurod1
|
neuronal differentiation 1 |
| chr10_-_28949111 | 1.27 |
ENSDART00000030138
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr23_-_7740845 | 1.27 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr7_+_49442972 | 1.26 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr20_-_45908435 | 1.26 |
ENSDART00000147637
|
fermt1
|
fermitin family member 1 |
| chr3_-_20807174 | 1.25 |
ENSDART00000159787
|
ndufa4l
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4, like |
| chr1_-_20131454 | 1.25 |
|
|
|
| chr18_+_43041488 | 1.24 |
|
|
|
| chr4_-_24298444 | 1.24 |
ENSDART00000077926
ENSDART00000128368 |
celf2
|
cugbp, Elav-like family member 2 |
| chr2_-_816669 | 1.24 |
ENSDART00000122732
|
foxc1a
|
forkhead box C1a |
| chr9_+_2756231 | 1.24 |
ENSDART00000123342
|
sp3a
|
sp3a transcription factor |
| chr10_-_43815475 | 1.24 |
ENSDART00000027392
|
mef2ca
|
myocyte enhancer factor 2ca |
| chr20_+_13998066 | 1.24 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr14_-_49115338 | 1.23 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr21_+_41684338 | 1.22 |
ENSDART00000169511
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr24_-_14432736 | 1.22 |
ENSDART00000135426
|
pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr9_-_33916709 | 1.22 |
ENSDART00000028225
|
mao
|
monoamine oxidase |
| chr24_+_21395671 | 1.21 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr8_-_31597252 | 1.21 |
ENSDART00000018886
|
ghra
|
growth hormone receptor a |
| chr4_+_57435 | 1.20 |
ENSDART00000169187
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr25_+_18460166 | 1.20 |
ENSDART00000073726
|
cav2
|
caveolin 2 |
| chr1_-_7456961 | 1.19 |
ENSDART00000152295
|
FAM83G
|
family with sequence similarity 83 member G |
| chr5_+_37053530 | 1.19 |
ENSDART00000161051
|
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr6_+_19795100 | 1.19 |
|
|
|
| chr6_-_2294751 | 1.19 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr19_+_17451381 | 1.18 |
ENSDART00000167602
|
spaca4l
|
sperm acrosome associated 4 like |
| chr8_+_29953399 | 1.18 |
ENSDART00000149372
ENSDART00000007640 |
ptch1
ptch1
|
patched 1 patched 1 |
| chr12_+_36716775 | 1.18 |
ENSDART00000048927
|
cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| KN149874v1_+_3991 | 1.18 |
|
|
|
| chr4_+_377829 | 1.18 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr7_-_7186964 | 1.18 |
ENSDART00000102620
|
six7
|
SIX homeobox 7 |
| chr6_-_23455346 | 1.18 |
ENSDART00000157527
ENSDART00000168882 ENSDART00000171384 |
ntn1a
|
netrin 1a |
| chr7_-_26153788 | 1.17 |
|
|
|
| chr19_+_22478256 | 1.17 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr20_+_34867305 | 1.17 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr3_-_58170499 | 1.17 |
ENSDART00000074272
|
im:6904045
|
im:6904045 |
| chr21_-_25486257 | 1.17 |
ENSDART00000110923
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr25_+_34629404 | 1.16 |
ENSDART00000156376
|
CR762436.2
|
Histone H3.2 |
| chr14_+_151136 | 1.16 |
ENSDART00000165766
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr8_-_18868986 | 1.16 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr25_-_13579869 | 1.15 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr4_-_9172622 | 1.15 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr23_+_17856053 | 1.14 |
ENSDART00000154427
|
CR381647.1
|
ENSDARG00000097211 |
| chr2_-_42011586 | 1.14 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr16_+_54906517 | 1.13 |
ENSDART00000126646
|
ENSDARG00000091079
|
ENSDARG00000091079 |
| chr12_+_34631310 | 1.13 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr22_+_20436009 | 1.12 |
ENSDART00000135984
|
onecut3a
|
one cut homeobox 3a |
| chr20_+_10502210 | 1.12 |
ENSDART00000155888
|
AL845550.1
|
ENSDARG00000097342 |
| chr25_+_27829693 | 1.11 |
ENSDART00000010325
|
fezf1
|
FEZ family zinc finger 1 |
| chr2_-_38242982 | 1.11 |
ENSDART00000040357
|
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr1_-_54319943 | 1.10 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr25_-_20886847 | 1.10 |
ENSDART00000110879
|
magi2b
|
membrane associated guanylate kinase, WW and PDZ domain containing 2b |
| chr7_-_73866202 | 1.10 |
|
|
|
| chr1_+_4557485 | 1.10 |
ENSDART00000052423
|
spry2
|
sprouty RTK signaling antagonist 2 |
| chr15_-_17074348 | 1.09 |
ENSDART00000156768
|
hip1
|
huntingtin interacting protein 1 |
| chr9_+_9133904 | 1.09 |
ENSDART00000165932
|
sik1
|
salt-inducible kinase 1 |
| chr1_-_53746151 | 1.09 |
ENSDART00000179565
|
CABZ01043826.1
|
ENSDARG00000108130 |
| chr23_+_19827551 | 1.09 |
ENSDART00000073442
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr7_-_33678058 | 1.08 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr15_+_43038684 | 1.08 |
ENSDART00000152222
|
acsl3a
|
acyl-CoA synthetase long-chain family member 3a |
| chr20_-_25726868 | 1.08 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr12_-_26760324 | 1.08 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr17_+_3842688 | 1.08 |
ENSDART00000170822
|
tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr7_+_73594790 | 1.08 |
ENSDART00000123081
|
zgc:173552
|
zgc:173552 |
| chr16_-_7321943 | 1.08 |
ENSDART00000149260
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| KN149681v1_-_46883 | 1.07 |
|
|
|
| chr3_+_25914076 | 1.07 |
ENSDART00000133523
|
hmgxb4a
|
HMG box domain containing 4a |
| chr8_+_3026357 | 1.07 |
ENSDART00000148020
ENSDART00000157599 |
lhx2b
|
LIM homeobox 2b |
| chr1_+_45063098 | 1.06 |
ENSDART00000084512
|
pkn1a
|
protein kinase N1a |
| chr16_+_16941228 | 1.06 |
ENSDART00000142155
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr16_+_24697776 | 1.06 |
|
|
|
| KN150207v1_-_1208 | 1.06 |
ENSDART00000171112
|
CABZ01112575.1
|
ENSDARG00000099354 |
| chr16_+_74373 | 1.05 |
|
|
|
| chr13_+_11305781 | 1.05 |
|
|
|
| chr20_+_53310252 | 1.05 |
ENSDART00000175125
|
CABZ01089116.1
|
ENSDARG00000106770 |
| chr23_-_19177660 | 1.05 |
ENSDART00000016901
|
pard6b
|
par-6 partitioning defective 6 homolog beta (C. elegans) |
| chr4_-_24298688 | 1.05 |
ENSDART00000077926
ENSDART00000128368 |
celf2
|
cugbp, Elav-like family member 2 |
| KN149874v1_+_3898 | 1.05 |
|
|
|
| chr3_+_26214852 | 1.05 |
ENSDART00000128613
|
rps15a
|
ribosomal protein S15a |
| chr22_+_528076 | 1.05 |
|
|
|
| chr3_+_24327586 | 1.04 |
ENSDART00000153551
|
cbx6b
|
chromobox homolog 6b |
| chr17_+_34854673 | 1.04 |
|
|
|
| chr8_+_160172 | 1.04 |
|
|
|
| chr16_-_41553835 | 1.03 |
ENSDART00000029492
|
cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0061114 | branching involved in pancreas morphogenesis(GO:0061114) pancreatic bud formation(GO:0061130) pancreas field specification(GO:0061131) |
| 0.8 | 2.4 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.7 | 2.2 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.7 | 3.4 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.6 | 1.9 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.6 | 3.1 | GO:1904325 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.6 | 1.8 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.6 | 2.3 | GO:0035908 | ventral aorta development(GO:0035908) pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.6 | 2.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.6 | 1.7 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.5 | 1.5 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.5 | 5.9 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.5 | 2.4 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.5 | 1.9 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.5 | 1.9 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.5 | 1.4 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.4 | 2.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.4 | 1.2 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.4 | 4.4 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.4 | 1.1 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.3 | 2.8 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.3 | 1.7 | GO:0071632 | optomotor response(GO:0071632) |
| 0.3 | 1.0 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.3 | 1.2 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.3 | 0.9 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.3 | 1.8 | GO:0034694 | response to prostaglandin(GO:0034694) |
| 0.3 | 1.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.3 | 2.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.3 | 2.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 1.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.3 | 0.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.3 | 1.1 | GO:0086065 | atrial cardiac muscle cell action potential(GO:0086014) cell-cell signaling involved in cardiac conduction(GO:0086019) atrial cardiac muscle cell to AV node cell signaling(GO:0086026) cell communication involved in cardiac conduction(GO:0086065) atrial cardiac muscle cell to AV node cell communication(GO:0086066) |
| 0.3 | 1.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 2.1 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.3 | 1.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 1.7 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 0.7 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) |
| 0.2 | 1.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 2.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.2 | 0.9 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.2 | 0.6 | GO:0014821 | ureteric bud development(GO:0001657) branching involved in ureteric bud morphogenesis(GO:0001658) mesonephros development(GO:0001823) phasic smooth muscle contraction(GO:0014821) ureteric bud morphogenesis(GO:0060675) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) mesonephric tubule morphogenesis(GO:0072171) |
| 0.2 | 1.3 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.2 | 0.8 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.2 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 2.7 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.2 | 0.9 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.2 | 3.0 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.2 | 0.7 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.2 | 1.2 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 0.5 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 0.3 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.2 | 4.2 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 1.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) |
| 0.2 | 0.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 0.3 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.2 | 1.3 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 0.5 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.2 | 1.3 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.2 | 1.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 0.5 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.2 | 0.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 0.5 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.2 | 1.1 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.2 | 0.8 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 0.9 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 1.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 0.5 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.2 | 2.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 2.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.9 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 3.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.8 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 1.0 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.1 | 1.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 1.0 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.1 | 1.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 2.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.9 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.1 | 1.0 | GO:0060579 | ventral spinal cord interneuron fate commitment(GO:0060579) |
| 0.1 | 2.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.5 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.1 | 0.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.3 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 1.2 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 1.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.3 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 1.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 0.9 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 0.5 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.4 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 7.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.5 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.3 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.1 | 0.9 | GO:0031128 | developmental induction(GO:0031128) Spemann organizer formation(GO:0060061) |
| 0.1 | 0.3 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.6 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.1 | 0.4 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.1 | 0.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 1.1 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 0.4 | GO:0031643 | positive regulation of myelination(GO:0031643) positive regulation of neurological system process(GO:0031646) |
| 0.1 | 0.8 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.6 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.4 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 1.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.3 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.1 | 1.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.8 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.2 | GO:0051963 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.7 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 1.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.4 | GO:0055017 | cardiac muscle tissue growth(GO:0055017) |
| 0.1 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.9 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.9 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 3.2 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.1 | 0.8 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.7 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.3 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 5.2 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 1.0 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 1.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.7 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 1.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.2 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 1.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 1.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 1.9 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 0.8 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.6 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.7 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 1.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 2.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 2.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.3 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.4 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 1.8 | GO:0010721 | negative regulation of cell development(GO:0010721) |
| 0.0 | 1.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.0 | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway(GO:0090100) |
| 0.0 | 0.3 | GO:0061036 | positive regulation of chondrocyte differentiation(GO:0032332) muscle cell fate commitment(GO:0042693) muscle cell fate specification(GO:0042694) positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.9 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 1.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.0 | 0.6 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.9 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.7 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 2.1 | GO:0099531 | neurotransmitter secretion(GO:0007269) presynaptic process involved in chemical synaptic transmission(GO:0099531) signal release from synapse(GO:0099643) |
| 0.0 | 2.7 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 1.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.4 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.4 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.4 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.9 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.1 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.6 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 3.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.7 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.6 | GO:0021761 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.6 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.7 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.3 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 3.6 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
| 0.0 | 1.5 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 2.3 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0048515 | spermatid development(GO:0007286) spermatid differentiation(GO:0048515) |
| 0.0 | 0.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 1.3 | GO:0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.5 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.0 | 0.7 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.7 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.3 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.5 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.3 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.5 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 0.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.1 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.3 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 2.5 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 1.2 | GO:0098916 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:0051311 | chiasma assembly(GO:0051026) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.2 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) ERBB signaling pathway(GO:0038127) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0001706 | endoderm formation(GO:0001706) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.4 | 1.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.4 | 2.0 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.3 | 0.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 1.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.3 | 1.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 0.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 1.0 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.2 | 4.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 1.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 0.9 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 0.7 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 7.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0044304 | main axon(GO:0044304) |
| 0.1 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 18.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 7.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.1 | 2.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 6.9 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.3 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.3 | GO:0044420 | basement membrane(GO:0005604) extracellular matrix component(GO:0044420) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 2.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 2.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 7.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 1.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 1.0 | GO:0022626 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 21.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.5 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.3 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0030120 | vesicle coat(GO:0030120) clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.9 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.0 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 2.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.6 | 1.8 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.6 | 1.7 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.5 | 1.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.5 | 3.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.4 | 3.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 2.2 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.4 | 1.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.4 | 1.3 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.4 | 3.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.4 | 1.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.4 | 1.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 5.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.4 | 1.1 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.3 | 1.7 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.3 | 1.0 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 1.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 1.2 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.3 | 1.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 1.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 1.4 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.3 | 0.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 0.7 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 2.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 0.7 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.2 | 0.9 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.2 | 2.2 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 2.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 1.2 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.2 | 0.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 0.9 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 1.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.2 | 0.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 0.7 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 3.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 0.5 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.2 | 0.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 1.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 2.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.7 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 1.1 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.9 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.3 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.1 | 0.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 1.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.5 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 3.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 0.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.6 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 2.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 2.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.4 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.4 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.3 | GO:0042910 | neurotransmitter transporter activity(GO:0005326) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.5 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 8.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 2.2 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0044620 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.0 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.5 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 50.9 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 1.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 1.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.9 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 2.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 4.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 1.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 1.6 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 6.2 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 3.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 3.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.5 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 2.7 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen(GO:0016705) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 2.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.7 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 1.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 2.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.3 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.5 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 4.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.3 | 0.6 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.2 | 1.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 2.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 4.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 1.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 3.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 3.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.5 | 2.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 2.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.4 | 5.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 1.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 1.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 1.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 6.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 1.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.7 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 0.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 2.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.0 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.1 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 0.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 3.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |