Project
DANIO-CODE
Navigation
Downloads
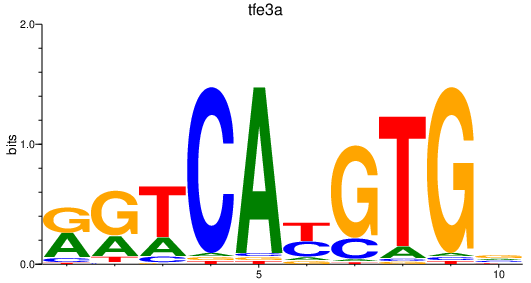
Results for tfe3a
Z-value: 0.84
Transcription factors associated with tfe3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfe3a
|
ENSDARG00000098903 | transcription factor binding to IGHM enhancer 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfe3a | dr10_dc_chr8_+_7739964_7740002 | -0.23 | 3.9e-01 | Click! |
Activity profile of tfe3a motif
Sorted Z-values of tfe3a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tfe3a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_32670368 | 1.62 |
ENSDART00000142449
|
ctsba
|
cathepsin Ba |
| chr12_-_2834771 | 1.39 |
ENSDART00000114854
ENSDART00000163759 |
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr13_+_50990946 | 1.32 |
|
|
|
| chr15_-_17163526 | 1.30 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr12_-_2834814 | 1.23 |
ENSDART00000114854
ENSDART00000163759 |
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr7_+_51520610 | 1.12 |
ENSDART00000174201
|
slc38a7
|
solute carrier family 38, member 7 |
| chr19_+_15536640 | 1.06 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr9_-_8318363 | 1.04 |
ENSDART00000139867
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr21_+_38024999 | 0.99 |
ENSDART00000142106
|
klf8
|
Kruppel-like factor 8 |
| chr2_-_57339717 | 0.96 |
ENSDART00000150034
|
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr6_+_135002 | 0.96 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| KN150699v1_-_15078 | 0.94 |
ENSDART00000159861
|
ENSDARG00000098739
|
ENSDARG00000098739 |
| chr23_-_31986679 | 0.92 |
ENSDART00000085054
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr23_-_22596648 | 0.92 |
ENSDART00000079019
|
spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr17_-_25313024 | 0.91 |
ENSDART00000082324
|
zpcx
|
zona pellucida protein C |
| chr19_-_42933592 | 0.90 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr10_-_44441390 | 0.89 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr14_+_30202682 | 0.88 |
ENSDART00000145336
|
her5
|
hairy-related 5 |
| chr13_-_17991273 | 0.87 |
ENSDART00000128748
|
fam21c
|
family with sequence similarity 21, member C |
| chr21_+_4375508 | 0.87 |
ENSDART00000043431
|
nup188
|
nucleoporin 188 |
| chr6_+_11162062 | 0.85 |
ENSDART00000151548
|
senp2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr15_+_1238560 | 0.80 |
ENSDART00000167760
ENSDART00000163827 |
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr16_-_4350251 | 0.80 |
|
|
|
| chr3_-_15325357 | 0.79 |
ENSDART00000139575
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr15_+_43682831 | 0.79 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr3_-_18655432 | 0.77 |
ENSDART00000034489
|
msrb1a
|
methionine sulfoxide reductase B1a |
| chr25_+_22222388 | 0.76 |
ENSDART00000154376
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr22_-_22337268 | 0.75 |
|
|
|
| chr8_-_52952141 | 0.74 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr13_+_46487890 | 0.74 |
|
|
|
| KN150563v1_-_10770 | 0.71 |
|
|
|
| chr9_-_56692366 | 0.71 |
|
|
|
| chr3_+_39426628 | 0.71 |
ENSDART00000137999
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr7_+_55217087 | 0.69 |
ENSDART00000149915
|
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr7_+_40866623 | 0.67 |
ENSDART00000052274
|
puf60b
|
poly-U binding splicing factor b |
| chr3_+_39426379 | 0.67 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr20_-_25732204 | 0.67 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr22_+_15317622 | 0.66 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| KN150421v1_+_14181 | 0.66 |
|
|
|
| chr3_-_32231069 | 0.66 |
ENSDART00000035545
|
prmt1
|
protein arginine methyltransferase 1 |
| chr24_-_30929990 | 0.66 |
|
|
|
| chr9_+_4382041 | 0.65 |
|
|
|
| chr4_-_7867294 | 0.65 |
ENSDART00000167943
|
nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr7_+_45747395 | 0.65 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr23_+_32102030 | 0.65 |
ENSDART00000145501
|
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr20_-_52002087 | 0.65 |
ENSDART00000041476
|
mia3
|
melanoma inhibitory activity family, member 3 |
| KN150563v1_-_10915 | 0.65 |
|
|
|
| chr24_-_30930178 | 0.65 |
|
|
|
| chr15_-_33669717 | 0.64 |
ENSDART00000161151
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr10_-_35205636 | 0.63 |
ENSDART00000131291
|
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr16_+_24045707 | 0.63 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr1_+_41644371 | 0.62 |
ENSDART00000143871
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr10_-_44441197 | 0.62 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr5_-_56539892 | 0.62 |
|
|
|
| chr16_+_4350448 | 0.61 |
|
|
|
| chr16_+_43468861 | 0.61 |
|
|
|
| chr12_+_30389706 | 0.60 |
|
|
|
| chr17_+_49995001 | 0.60 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr17_+_23291331 | 0.59 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr19_-_808376 | 0.58 |
ENSDART00000140119
|
glmp
|
glycosylated lysosomal membrane protein |
| chr23_+_38313746 | 0.58 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr4_+_17666992 | 0.57 |
ENSDART00000066999
|
ccdc53
|
coiled-coil domain containing 53 |
| chr15_-_14102102 | 0.57 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr7_+_1443102 | 0.57 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr15_-_33669618 | 0.57 |
ENSDART00000161151
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr10_-_24754849 | 0.56 |
ENSDART00000148582
|
smpd1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr16_+_33209956 | 0.56 |
ENSDART00000101943
|
rragca
|
Ras-related GTP binding Ca |
| chr16_+_20690902 | 0.56 |
ENSDART00000078984
|
cpvl
|
carboxypeptidase, vitellogenic-like |
| chr23_+_26153043 | 0.56 |
ENSDART00000054013
|
atp6ap1b
|
ATPase, H+ transporting, lysosomal accessory protein 1b |
| chr1_-_39141284 | 0.55 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr14_-_46608307 | 0.55 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr8_-_44247277 | 0.55 |
|
|
|
| chr18_+_14725283 | 0.54 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr21_+_7737161 | 0.54 |
|
|
|
| chr18_-_16927749 | 0.54 |
ENSDART00000100105
ENSDART00000163400 |
swap70b
|
SWAP switching B-cell complex 70b subunit 70b |
| chr20_+_26005068 | 0.53 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr15_-_25025251 | 0.53 |
ENSDART00000109990
|
abhd15a
|
abhydrolase domain containing 15a |
| chr17_+_17745119 | 0.53 |
ENSDART00000138189
ENSDART00000105013 |
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr23_+_10870124 | 0.52 |
ENSDART00000035693
|
ppp4r2a
|
protein phosphatase 4, regulatory subunit 2a |
| chr20_+_26495546 | 0.51 |
ENSDART00000036942
|
zbtb2b
|
zinc finger and BTB domain containing 2b |
| chr3_+_4123775 | 0.51 |
ENSDART00000056035
|
ENSDARG00000038398
|
ENSDARG00000038398 |
| chr11_-_25019298 | 0.51 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr16_-_5243402 | 0.50 |
ENSDART00000131876
|
ttk
|
ttk protein kinase |
| chr21_-_3548863 | 0.50 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr7_+_27184121 | 0.50 |
ENSDART00000148417
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr15_+_35081979 | 0.50 |
ENSDART00000131182
|
zgc:66024
|
zgc:66024 |
| chr20_-_51287350 | 0.50 |
ENSDART00000023488
|
atp6v1d
|
ATPase, H+ transporting, lysosomal V1 subunit D |
| chr18_+_27095398 | 0.50 |
ENSDART00000125326
ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr14_+_21373857 | 0.50 |
ENSDART00000079649
ENSDART00000165795 |
ndufs8b
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8b |
| chr25_-_35819452 | 0.49 |
ENSDART00000064400
|
si:ch211-113a14.24
|
si:ch211-113a14.24 |
| chr15_+_1569674 | 0.49 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr14_-_17270022 | 0.49 |
ENSDART00000123145
|
rnf4
|
ring finger protein 4 |
| chr11_-_14044356 | 0.49 |
ENSDART00000085158
|
tmem259
|
transmembrane protein 259 |
| chr5_+_50266345 | 0.48 |
ENSDART00000092938
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr3_-_15325178 | 0.48 |
ENSDART00000139575
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr3_-_32231192 | 0.48 |
ENSDART00000012630
|
prmt1
|
protein arginine methyltransferase 1 |
| chr5_-_54091790 | 0.48 |
ENSDART00000150070
|
ccnb1
|
cyclin B1 |
| chr8_-_52952226 | 0.47 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr25_+_2968769 | 0.47 |
|
|
|
| chr3_+_14314116 | 0.47 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr21_+_10692781 | 0.47 |
|
|
|
| chr3_+_31530419 | 0.47 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr13_+_25374720 | 0.46 |
ENSDART00000140634
|
inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr10_+_32107014 | 0.46 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr25_-_19510499 | 0.46 |
ENSDART00000166824
|
gtse1
|
G-2 and S-phase expressed 1 |
| chr13_-_2296041 | 0.46 |
ENSDART00000126697
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr17_+_24790812 | 0.46 |
ENSDART00000082251
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr2_+_25281122 | 0.46 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr17_+_30826890 | 0.46 |
ENSDART00000149600
|
tpp1
|
tripeptidyl peptidase I |
| chr5_-_23296575 | 0.45 |
ENSDART00000134184
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr25_-_24105014 | 0.45 |
ENSDART00000048507
|
uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr9_-_56692281 | 0.45 |
|
|
|
| chr13_-_35924447 | 0.44 |
ENSDART00000134955
|
lgmn
|
legumain |
| chr10_+_5267937 | 0.44 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr11_-_38856849 | 0.44 |
ENSDART00000113185
|
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr7_-_41532268 | 0.44 |
ENSDART00000016105
ENSDART00000138800 |
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr16_-_31664756 | 0.44 |
ENSDART00000176939
|
phf20l1
|
PHD finger protein 20-like 1 |
| chr3_-_42949449 | 0.44 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr3_+_42217187 | 0.44 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr24_-_20946041 | 0.44 |
ENSDART00000140786
|
qtrtd1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr13_-_31557441 | 0.43 |
ENSDART00000085989
|
trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) |
| chr14_-_35513309 | 0.43 |
ENSDART00000173343
|
pdgfc
|
platelet derived growth factor c |
| chr4_-_765957 | 0.43 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr23_+_34021303 | 0.43 |
ENSDART00000136104
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr19_-_2148053 | 0.42 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr7_-_19713757 | 0.42 |
|
|
|
| chr5_-_9036049 | 0.42 |
ENSDART00000160079
|
gak
|
cyclin G associated kinase |
| chr18_-_1745485 | 0.42 |
|
|
|
| chr16_+_24045774 | 0.42 |
ENSDART00000133484
|
apoeb
|
apolipoprotein Eb |
| chr6_+_54672873 | 0.42 |
ENSDART00000074602
|
smpd2b
|
sphingomyelin phosphodiesterase 2b, neutral membrane (neutral sphingomyelinase) |
| chr3_-_22240424 | 0.42 |
|
|
|
| chr24_-_30929960 | 0.42 |
|
|
|
| chr21_+_38025480 | 0.42 |
ENSDART00000142106
|
klf8
|
Kruppel-like factor 8 |
| chr24_+_40321763 | 0.42 |
|
|
|
| chr17_+_7377230 | 0.41 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr14_+_15982295 | 0.41 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr13_-_4863766 | 0.41 |
ENSDART00000159679
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr23_+_20586745 | 0.41 |
ENSDART00000157522
|
dpm1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr3_+_42217236 | 0.41 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr10_-_35205597 | 0.41 |
ENSDART00000131291
|
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr11_+_38858311 | 0.41 |
ENSDART00000155746
|
cdc42
|
cell division cycle 42 |
| chr18_-_26131468 | 0.41 |
|
|
|
| chr4_+_18427325 | 0.41 |
|
|
|
| chr7_+_41532360 | 0.41 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr14_-_47387481 | 0.40 |
ENSDART00000031498
|
ccna2
|
cyclin A2 |
| chr7_+_25554355 | 0.40 |
ENSDART00000149835
|
mtm1
|
myotubularin 1 |
| chr16_+_19223683 | 0.40 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr23_-_2957262 | 0.40 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr6_+_131451 | 0.40 |
ENSDART00000166361
|
fdx1l
|
ferredoxin 1-like |
| chr20_-_223222 | 0.40 |
ENSDART00000104790
|
znf292b
|
zinc finger protein 292b |
| chr2_-_23330863 | 0.39 |
|
|
|
| chr23_-_19125310 | 0.39 |
ENSDART00000140866
|
arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr12_+_33219727 | 0.39 |
ENSDART00000021491
|
csnk1db
|
casein kinase 1, delta b |
| chr11_+_44678488 | 0.39 |
ENSDART00000163408
|
si:dkey-93h22.8
|
si:dkey-93h22.8 |
| chr15_-_33669589 | 0.39 |
ENSDART00000161151
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr21_-_19883182 | 0.39 |
ENSDART00000065670
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr1_-_28257339 | 0.39 |
ENSDART00000148331
|
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr23_-_18105560 | 0.39 |
|
|
|
| chr9_+_42355513 | 0.39 |
ENSDART00000142888
|
lrrc3
|
leucine rich repeat containing 3 |
| chr24_+_35296642 | 0.38 |
ENSDART00000172652
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr14_+_20927461 | 0.38 |
ENSDART00000160576
|
si:ch211-175m2.4
|
si:ch211-175m2.4 |
| chr14_+_5528229 | 0.38 |
ENSDART00000140260
|
aup1
|
ancient ubiquitous protein 1 |
| chr8_-_47339228 | 0.38 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr21_+_6289363 | 0.38 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr8_-_29842489 | 0.37 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr17_-_44326992 | 0.37 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr8_-_16689736 | 0.37 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr17_-_20267580 | 0.37 |
ENSDART00000078703
|
add3b
|
adducin 3 (gamma) b |
| chr5_-_60664244 | 0.37 |
ENSDART00000136553
|
si:ch211-209a2.2
|
si:ch211-209a2.2 |
| chr3_-_15992035 | 0.37 |
ENSDART00000016616
|
arl5c
|
ADP-ribosylation factor-like 5C |
| chr4_+_5332964 | 0.37 |
ENSDART00000123375
|
zgc:113263
|
zgc:113263 |
| chr13_-_29571309 | 0.37 |
ENSDART00000139773
|
CT025909.1
|
ENSDARG00000093614 |
| chr24_+_35296732 | 0.37 |
ENSDART00000172652
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr1_-_7882397 | 0.36 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr23_+_26152909 | 0.36 |
ENSDART00000129617
|
atp6ap1b
|
ATPase, H+ transporting, lysosomal accessory protein 1b |
| chr19_+_365865 | 0.36 |
ENSDART00000093383
|
vps72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr6_+_135255 | 0.36 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr17_+_24791024 | 0.36 |
ENSDART00000130871
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr8_-_16614882 | 0.36 |
ENSDART00000135319
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr10_-_15921335 | 0.36 |
ENSDART00000136789
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr4_-_11126782 | 0.36 |
ENSDART00000150419
|
BX784029.1
|
ENSDARG00000096039 |
| chr20_-_25732091 | 0.36 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr17_+_25313170 | 0.36 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr13_+_25374865 | 0.35 |
ENSDART00000140634
|
inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr13_-_2296283 | 0.35 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr8_-_20213672 | 0.35 |
ENSDART00000136911
|
acer1
|
alkaline ceramidase 1 |
| chr15_-_16241412 | 0.35 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr14_-_21779536 | 0.35 |
|
|
|
| chr18_+_16144493 | 0.35 |
ENSDART00000080423
|
ctsd
|
cathepsin D |
| chr19_-_27239547 | 0.35 |
ENSDART00000149132
|
neu1
|
neuraminidase 1 |
| chr5_-_32290516 | 0.35 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr24_-_36382704 | 0.35 |
ENSDART00000154858
|
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr3_-_25290808 | 0.35 |
ENSDART00000154200
|
bptf
|
bromodomain PHD finger transcription factor |
| chr7_+_28905795 | 0.35 |
ENSDART00000146655
|
aph1b
|
APH1B gamma secretase subunit |
| chr15_+_43682994 | 0.35 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr22_+_21491685 | 0.35 |
ENSDART00000136374
|
mier2
|
mesoderm induction early response 1, family member 2 |
| chr16_+_14326410 | 0.34 |
ENSDART00000113093
|
gba
|
glucosidase, beta, acid |
| chr20_-_16949608 | 0.34 |
ENSDART00000027582
|
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr24_-_42132495 | 0.34 |
|
|
|
| chr24_-_30930283 | 0.34 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.4 | 1.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.3 | 1.7 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.3 | 1.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.3 | 0.8 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 1.7 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.2 | 0.9 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.2 | 0.6 | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process(GO:1902633) 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process(GO:1902635) |
| 0.2 | 0.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 0.5 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.2 | 0.5 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.2 | 0.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 0.5 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.2 | 0.6 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.2 | 0.6 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.4 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 0.8 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 1.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.6 | GO:0006226 | dUMP biosynthetic process(GO:0006226) dUMP metabolic process(GO:0046078) |
| 0.1 | 0.8 | GO:1902423 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) regulation of mitotic attachment of spindle microtubules to kinetochore(GO:1902423) positive regulation of attachment of mitotic spindle microtubules to kinetochore(GO:1902425) RNA localization to chromatin(GO:1990280) |
| 0.1 | 0.7 | GO:0032516 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 1.4 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.9 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.3 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.3 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.1 | 0.4 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.3 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.1 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.3 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.1 | 0.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.3 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.4 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.6 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 1.3 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 0.4 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.1 | 0.3 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 0.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.7 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.4 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.1 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.5 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.1 | GO:2000009 | negative regulation of cellular protein localization(GO:1903828) regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.2 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.6 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.3 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.3 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 0.2 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 1.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.8 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.6 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 0.2 | GO:0061011 | meiotic DNA double-strand break processing(GO:0000706) hepatic duct development(GO:0061011) |
| 0.1 | 0.2 | GO:0010662 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.1 | 0.2 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) male sex differentiation(GO:0046661) |
| 0.1 | 0.2 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.2 | GO:0009092 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.1 | 1.7 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 0.8 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.2 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.1 | 0.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 1.8 | GO:0048599 | oocyte development(GO:0048599) |
| 0.1 | 0.9 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.5 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.1 | 2.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.3 | GO:0034341 | monocyte chemotaxis(GO:0002548) response to interferon-gamma(GO:0034341) cellular response to interferon-gamma(GO:0071346) |
| 0.1 | 0.3 | GO:0046386 | deoxyribonucleotide catabolic process(GO:0009264) deoxyribose phosphate catabolic process(GO:0046386) |
| 0.1 | 1.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.2 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.5 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.1 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.7 | GO:0006684 | sphingomyelin metabolic process(GO:0006684) |
| 0.0 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.6 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.0 | 0.2 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.6 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.6 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.6 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.8 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.6 | GO:0006000 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.0 | 0.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.6 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 1.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.3 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.6 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.1 | GO:0008542 | associative learning(GO:0008306) visual learning(GO:0008542) mammillary axonal complex development(GO:0061373) mammillothalamic axonal tract development(GO:0061374) corpora quadrigemina development(GO:0061378) inferior colliculus development(GO:0061379) cell migration in diencephalon(GO:0061381) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 2.0 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.3 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 1.2 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 1.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.4 | GO:0045494 | Golgi to plasma membrane transport(GO:0006893) photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.1 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.3 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.0 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.0 | 0.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.2 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.1 | GO:0007135 | meiosis II(GO:0007135) meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.0 | 0.5 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.2 | GO:1903321 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 0.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 1.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 0.8 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.2 | 1.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.4 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.8 | GO:0000803 | sex chromosome(GO:0000803) inactive sex chromosome(GO:0098577) |
| 0.1 | 0.9 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.6 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 1.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.6 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 1.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 0.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.4 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0030666 | endocytic vesicle membrane(GO:0030666) phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 5.9 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.3 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.4 | 1.3 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.4 | 1.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 0.8 | GO:0016595 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.3 | 0.8 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 1.1 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 0.7 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 0.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 1.0 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 0.9 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.5 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 0.8 | GO:0003948 | N4-(beta-N-acetylglucosaminyl)-L-asparaginase activity(GO:0003948) |
| 0.2 | 0.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.8 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.1 | 0.8 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.9 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.8 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.3 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.3 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 1.3 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 0.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.4 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.1 | 0.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.3 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 2.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.3 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.1 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.2 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 3.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.2 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) steroid hormone receptor binding(GO:0035258) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 2.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0016894 | endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 3'-phosphomonoesters(GO:0016894) |
| 0.0 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.3 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.1 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |