Project
DANIO-CODE
Navigation
Downloads
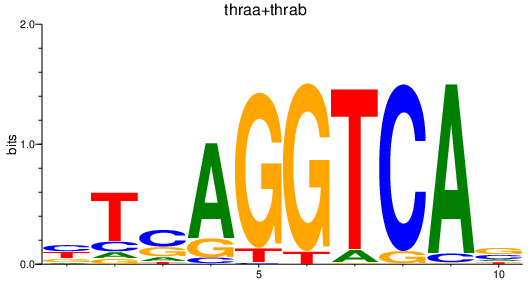
Results for thraa+thrab
Z-value: 1.59
Transcription factors associated with thraa+thrab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
thraa
|
ENSDARG00000000151 | thyroid hormone receptor alpha a |
|
thrab
|
ENSDARG00000052654 | thyroid hormone receptor alpha b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| thrab | dr10_dc_chr12_-_21918058_21918109 | 0.71 | 2.0e-03 | Click! |
| thraa | dr10_dc_chr3_-_34623996_34624121 | -0.54 | 3.0e-02 | Click! |
Activity profile of thraa+thrab motif
Sorted Z-values of thraa+thrab motif
Network of associatons between targets according to the STRING database.
First level regulatory network of thraa+thrab
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_41347831 | 4.25 |
ENSDART00000174304
|
plxdc2
|
plexin domain containing 2 |
| chr6_-_13654186 | 4.03 |
ENSDART00000150102
ENSDART00000041269 |
cryba2a
|
crystallin, beta A2a |
| chr15_+_37204488 | 3.48 |
ENSDART00000157762
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr12_-_3098744 | 3.34 |
ENSDART00000015092
|
col1a1b
|
collagen, type I, alpha 1b |
| chr18_-_15404998 | 3.32 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr7_+_19765237 | 3.31 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr21_+_39140701 | 3.22 |
ENSDART00000075992
|
cryba1b
|
crystallin, beta A1b |
| chr7_-_16776026 | 3.18 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr23_-_26150495 | 3.07 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr23_+_44817648 | 3.05 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr24_-_39722595 | 3.04 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr10_+_6316944 | 3.03 |
ENSDART00000162428
|
tpm2
|
tropomyosin 2 (beta) |
| chr2_-_28446615 | 2.91 |
ENSDART00000179495
|
cdh6
|
cadherin 6 |
| chr19_-_24859287 | 2.89 |
ENSDART00000163763
|
thbs3b
|
thrombospondin 3b |
| chr9_-_22461196 | 2.86 |
ENSDART00000135190
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr10_-_44180588 | 2.81 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr6_+_41466199 | 2.73 |
ENSDART00000177439
|
twf2a
|
twinfilin actin-binding protein 2a |
| chr13_-_37144297 | 2.70 |
|
|
|
| chr9_-_22318918 | 2.64 |
ENSDART00000124272
|
crygm2d17
|
crystallin, gamma M2d17 |
| chr20_+_45989418 | 2.64 |
ENSDART00000131169
|
bmp2b
|
bone morphogenetic protein 2b |
| chr18_-_15405161 | 2.62 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr11_+_25022419 | 2.57 |
|
|
|
| chr9_-_22470612 | 2.57 |
ENSDART00000123763
|
crygm2d3
|
crystallin, gamma M2d3 |
| chr1_-_44476084 | 2.56 |
ENSDART00000034549
|
zgc:111983
|
zgc:111983 |
| chr17_+_381715 | 2.55 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr3_-_29846530 | 2.55 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr15_+_24708696 | 2.54 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr13_-_30515486 | 2.51 |
ENSDART00000139607
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr9_-_22394674 | 2.51 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr19_+_3713027 | 2.44 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr16_+_53238110 | 2.44 |
ENSDART00000102170
|
CABZ01053976.1
|
ENSDARG00000069929 |
| chr19_-_47897712 | 2.42 |
|
|
|
| chr6_+_59788008 | 2.38 |
|
|
|
| chr4_-_1346961 | 2.34 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr16_+_33702010 | 2.33 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr11_-_29891067 | 2.31 |
ENSDART00000172106
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr5_+_26925238 | 2.31 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr5_+_26840147 | 2.31 |
ENSDART00000064701
|
loxl2b
|
lysyl oxidase-like 2b |
| chr2_+_33399405 | 2.27 |
ENSDART00000137207
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr20_+_34679181 | 2.26 |
|
|
|
| chr9_-_22434581 | 2.26 |
ENSDART00000114943
|
crygm2d4
|
crystallin, gamma M2d4 |
| chr9_-_22488422 | 2.24 |
ENSDART00000124600
|
crygm2d21
|
crystallin, gamma M2d21 |
| chr3_-_55995924 | 2.20 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr4_-_25282256 | 2.18 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr10_-_22881453 | 2.17 |
|
|
|
| chr9_-_22528568 | 2.15 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr8_+_28240085 | 2.15 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr20_-_17006936 | 2.10 |
ENSDART00000012859
|
psma6b
|
proteasome subunit alpha 6b |
| chr7_+_28862183 | 2.07 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr1_-_20218263 | 2.04 |
ENSDART00000078271
|
cpe
|
carboxypeptidase E |
| chr3_+_33168814 | 2.02 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr3_-_18426055 | 2.00 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr13_-_31304614 | 1.96 |
|
|
|
| chr13_-_39821399 | 1.95 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr18_-_8697347 | 1.93 |
ENSDART00000093131
|
FRMD4A
|
FERM domain containing 4A |
| chr21_-_13571753 | 1.92 |
ENSDART00000065819
|
clic3
|
chloride intracellular channel 3 |
| chr19_+_42491275 | 1.91 |
|
|
|
| chr25_-_325847 | 1.90 |
ENSDART00000059514
|
prickle1a
|
prickle homolog 1a |
| chr5_-_30015572 | 1.89 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr16_-_44575942 | 1.88 |
|
|
|
| chr7_-_29811734 | 1.88 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr10_-_44441481 | 1.86 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr4_-_25282161 | 1.84 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr19_-_39048324 | 1.82 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr18_-_11216447 | 1.81 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr19_-_7069920 | 1.79 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr19_+_3925188 | 1.77 |
|
|
|
| chr1_+_38834751 | 1.76 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr3_-_13942505 | 1.74 |
|
|
|
| chr5_-_15774816 | 1.71 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr6_+_30441419 | 1.70 |
|
|
|
| chr25_+_5695630 | 1.70 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr21_+_25651712 | 1.68 |
ENSDART00000135123
|
si:dkey-17e16.17
|
si:dkey-17e16.17 |
| chr25_+_35901081 | 1.66 |
ENSDART00000042271
ENSDART00000158027 |
irx3b
|
iroquois homeobox 3b |
| chr23_+_6298911 | 1.60 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| KN150131v1_+_59098 | 1.60 |
ENSDART00000175590
|
CABZ01064941.1
|
ENSDARG00000106550 |
| chr7_-_26036887 | 1.60 |
ENSDART00000146935
|
zgc:77439
|
zgc:77439 |
| chr5_+_26925392 | 1.59 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr10_-_39210658 | 1.59 |
ENSDART00000150193
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr14_-_32877752 | 1.59 |
ENSDART00000163046
|
si:dkey-31j3.11
|
si:dkey-31j3.11 |
| chr15_-_10345337 | 1.57 |
|
|
|
| chr9_-_56692366 | 1.53 |
|
|
|
| chr24_+_14302079 | 1.50 |
ENSDART00000168059
|
prdm14
|
PR domain containing 14 |
| chr9_+_907908 | 1.50 |
ENSDART00000144114
|
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr18_-_15963983 | 1.47 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr9_-_31714290 | 1.46 |
ENSDART00000127214
|
tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr1_-_19001718 | 1.46 |
ENSDART00000143303
|
BX323590.2
|
ENSDARG00000095388 |
| chr11_-_6004509 | 1.46 |
ENSDART00000108628
|
ano8b
|
anoctamin 8b |
| chr17_+_33423057 | 1.44 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr25_-_28691412 | 1.44 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr9_-_22324699 | 1.42 |
ENSDART00000175417
ENSDART00000101902 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr24_-_42038250 | 1.42 |
ENSDART00000171380
|
top1mt
|
topoisomerase (DNA) I, mitochondrial |
| chr16_+_5778650 | 1.42 |
ENSDART00000131575
|
zgc:158689
|
zgc:158689 |
| chr14_-_6822151 | 1.41 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr13_-_16126849 | 1.39 |
ENSDART00000079745
|
zgc:110045
|
zgc:110045 |
| chr24_-_16995194 | 1.38 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr2_+_33907298 | 1.38 |
|
|
|
| chr5_+_37861949 | 1.38 |
ENSDART00000144425
|
gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr11_+_6809190 | 1.36 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr16_+_33701759 | 1.36 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr16_-_33105847 | 1.36 |
|
|
|
| chr4_+_26507297 | 1.35 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr23_-_7892862 | 1.33 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr13_-_14978365 | 1.33 |
|
|
|
| chr11_-_1374026 | 1.31 |
ENSDART00000172953
|
rpl29
|
ribosomal protein L29 |
| chr14_-_47688669 | 1.30 |
ENSDART00000113285
|
tapt1a
|
transmembrane anterior posterior transformation 1a |
| chr21_-_37404394 | 1.30 |
ENSDART00000129439
|
BX649250.1
|
ENSDARG00000091187 |
| chr22_+_26423320 | 1.30 |
ENSDART00000044085
|
ENSDARG00000034211
|
ENSDARG00000034211 |
| chr22_+_26616977 | 1.29 |
ENSDART00000171972
ENSDART00000083351 |
ENSDARG00000034211
|
ENSDARG00000034211 |
| chr24_-_40968409 | 1.29 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr19_+_4996328 | 1.28 |
ENSDART00000165082
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr18_-_48498261 | 1.28 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr24_-_16994956 | 1.26 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr1_+_28293053 | 1.26 |
ENSDART00000075539
|
cryaa
|
crystallin, alpha A |
| chr8_-_2375664 | 1.25 |
ENSDART00000144017
|
vdac3
|
voltage-dependent anion channel 3 |
| chr25_+_36872560 | 1.25 |
ENSDART00000163178
|
slc10a3
|
solute carrier family 10, member 3 |
| chr19_-_22983970 | 1.25 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr4_-_71353374 | 1.25 |
|
|
|
| chr1_+_1964902 | 1.23 |
ENSDART00000138396
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr15_+_1238615 | 1.23 |
ENSDART00000167760
ENSDART00000163827 |
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr17_+_33422832 | 1.22 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr15_+_29823641 | 1.22 |
|
|
|
| chr15_-_2762950 | 1.21 |
ENSDART00000108856
|
si:ch211-235m3.5
|
si:ch211-235m3.5 |
| chr15_+_9321225 | 1.20 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr13_-_49695322 | 1.18 |
|
|
|
| chr19_-_7069850 | 1.17 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr7_+_38008155 | 1.15 |
ENSDART00000169927
|
lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr10_+_8670564 | 1.13 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr8_-_6875110 | 1.11 |
ENSDART00000014915
|
asb6
|
ankyrin repeat and SOCS box containing 6 |
| chr18_-_16934961 | 1.10 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr6_-_32000976 | 1.08 |
ENSDART00000132280
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr11_-_11487856 | 1.07 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr13_+_51005155 | 1.06 |
|
|
|
| chr3_-_29846789 | 1.06 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr12_-_29190576 | 1.06 |
ENSDART00000153458
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr23_-_3760604 | 1.05 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr17_+_8874210 | 1.04 |
|
|
|
| chr16_+_6693574 | 1.04 |
|
|
|
| chr14_+_15698814 | 1.04 |
|
|
|
| chr23_-_45356039 | 1.03 |
ENSDART00000067630
|
ENSDARG00000039901
|
ENSDARG00000039901 |
| chr10_+_39211384 | 1.01 |
|
|
|
| chr22_-_29387056 | 0.99 |
ENSDART00000121599
|
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr8_-_20213672 | 0.98 |
ENSDART00000136911
|
acer1
|
alkaline ceramidase 1 |
| chr5_+_14850708 | 0.98 |
ENSDART00000140990
ENSDART00000137287 |
pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr6_+_60190849 | 0.96 |
|
|
|
| chr13_-_36577270 | 0.95 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr9_-_48202667 | 0.94 |
|
|
|
| chr25_+_5695588 | 0.94 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr14_+_33073643 | 0.93 |
ENSDART00000052780
|
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr16_+_24046789 | 0.92 |
|
|
|
| chr6_+_7257078 | 0.92 |
ENSDART00000049339
|
sox21a
|
SRY (sex determining region Y)-box 21a |
| chr4_+_7499654 | 0.92 |
ENSDART00000158999
|
tnnt2e
|
troponin T2e, cardiac |
| chr10_-_39211281 | 0.92 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr2_+_24718602 | 0.92 |
ENSDART00000022379
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr10_+_29966381 | 0.92 |
ENSDART00000141549
|
hspa8
|
heat shock protein 8 |
| chr19_-_39048402 | 0.91 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr20_+_316191 | 0.91 |
ENSDART00000104807
|
si:dkey-119m7.4
|
si:dkey-119m7.4 |
| chr7_+_30803603 | 0.88 |
ENSDART00000173805
|
tjp1a
|
tight junction protein 1a |
| chr23_+_24161550 | 0.88 |
ENSDART00000145159
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr11_+_23012136 | 0.87 |
ENSDART00000110152
|
csf1a
|
colony stimulating factor 1a (macrophage) |
| chr1_+_23093114 | 0.86 |
|
|
|
| chr19_-_22984081 | 0.82 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr16_-_31835463 | 0.82 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr5_-_21520111 | 0.82 |
|
|
|
| chr7_+_26980284 | 0.80 |
ENSDART00000173962
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr23_-_4919504 | 0.80 |
ENSDART00000133701
|
slc6a1a
|
solute carrier family 6 (neurotransmitter transporter), member 1a |
| chr22_-_62082 | 0.80 |
ENSDART00000166010
ENSDART00000125700 |
mrpl20
|
mitochondrial ribosomal protein L20 |
| chr3_-_34417082 | 0.78 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr1_-_15814788 | 0.78 |
ENSDART00000169081
|
mtmr7b
|
myotubularin related protein 7b |
| chr8_-_46903869 | 0.77 |
ENSDART00000161462
|
acot7
|
acyl-CoA thioesterase 7 |
| chr3_-_19346061 | 0.77 |
ENSDART00000162248
|
CU571315.3
|
ENSDARG00000101990 |
| chr17_-_15141309 | 0.77 |
ENSDART00000103405
|
gch1
|
GTP cyclohydrolase 1 |
| chr13_-_30515304 | 0.76 |
ENSDART00000139607
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr23_-_39858232 | 0.75 |
ENSDART00000160853
|
minos1
|
mitochondrial inner membrane organizing system 1 |
| chr15_+_43683110 | 0.74 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr10_-_14587464 | 0.71 |
ENSDART00000057865
|
ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr13_-_29294141 | 0.71 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr5_+_52198613 | 0.70 |
ENSDART00000162459
|
scarb2a
|
scavenger receptor class B, member 2a |
| chr8_+_24725855 | 0.70 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr16_+_68014 | 0.70 |
ENSDART00000159652
|
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr25_-_15117843 | 0.69 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr2_-_30005139 | 0.69 |
ENSDART00000146968
|
cnpy1
|
canopy1 |
| chr16_-_5821719 | 0.68 |
ENSDART00000136655
|
ndufa3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 |
| chr5_-_32686309 | 0.66 |
ENSDART00000117288
|
SNORD24
|
Small nucleolar RNA SNORD24 |
| chr5_-_56534037 | 0.65 |
ENSDART00000171252
|
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr21_+_35292807 | 0.64 |
ENSDART00000130626
|
rars
|
arginyl-tRNA synthetase |
| chr16_+_54001592 | 0.64 |
|
|
|
| chr23_-_4175790 | 0.64 |
ENSDART00000109807
|
ENSDARG00000076299
|
ENSDARG00000076299 |
| chr14_-_32877816 | 0.64 |
ENSDART00000163046
|
si:dkey-31j3.11
|
si:dkey-31j3.11 |
| chr20_+_36720896 | 0.64 |
ENSDART00000062895
|
srp9
|
signal recognition particle 9 |
| chr14_-_34431408 | 0.64 |
ENSDART00000141157
ENSDART00000160598 ENSDART00000150413 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr10_+_29966252 | 0.62 |
ENSDART00000141549
|
hspa8
|
heat shock protein 8 |
| chr1_-_1254253 | 0.61 |
ENSDART00000130697
|
runx1
|
runt-related transcription factor 1 |
| chr9_+_41776713 | 0.61 |
|
|
|
| chr14_-_2179471 | 0.59 |
ENSDART00000157401
|
pcdh2g1
|
protocadherin 2 gamma 1 |
| chr14_-_38734382 | 0.59 |
ENSDART00000030509
|
glra4a
|
glycine receptor, alpha 4a |
| chr24_+_33697332 | 0.58 |
|
|
|
| chr4_+_8007802 | 0.58 |
ENSDART00000014036
|
optn
|
optineurin |
| chr9_-_30560440 | 0.58 |
ENSDART00000079068
|
asb11
|
ankyrin repeat and SOCS box containing 11 |
| chr9_-_34690993 | 0.58 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 1.0 | 3.1 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.9 | 2.6 | GO:0061130 | branching involved in pancreas morphogenesis(GO:0061114) pancreatic bud formation(GO:0061130) pancreas field specification(GO:0061131) |
| 0.8 | 2.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.6 | 1.9 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.6 | 1.8 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.6 | 2.3 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.5 | 2.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 2.2 | GO:0048901 | anterior lateral line neuromast development(GO:0048901) anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.4 | 1.3 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.4 | 27.7 | GO:0007601 | visual perception(GO:0007601) |
| 0.3 | 1.4 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.3 | 1.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 1.7 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.3 | 1.9 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.3 | 0.9 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 0.8 | GO:1990092 | calcium-dependent self proteolysis(GO:1990092) |
| 0.3 | 1.5 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.2 | 0.7 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 1.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 0.6 | GO:0030852 | regulation of collagen metabolic process(GO:0010712) regulation of granulocyte differentiation(GO:0030852) plasminogen activation(GO:0031639) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) regulation of neutrophil differentiation(GO:0045658) |
| 0.2 | 0.8 | GO:0061036 | positive regulation of chondrocyte differentiation(GO:0032332) positive regulation of cartilage development(GO:0061036) |
| 0.2 | 2.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.8 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.2 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.4 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 1.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 0.7 | GO:0072149 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.2 | 0.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.9 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 2.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 2.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:0002762 | negative regulation of myeloid leukocyte differentiation(GO:0002762) |
| 0.1 | 1.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 3.8 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.8 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 1.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 2.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 2.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.0 | GO:0097320 | membrane tubulation(GO:0097320) positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.5 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.6 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 1.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 1.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 1.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.2 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 0.6 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.1 | 1.2 | GO:0047496 | vesicle transport along microtubule(GO:0047496) |
| 0.0 | 3.2 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 2.6 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 2.3 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 4.0 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.0 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.5 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 1.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.5 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.7 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 2.0 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 1.1 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.2 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.4 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.1 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 1.4 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 0.9 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 2.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 2.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 4.0 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 2.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 4.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 3.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 1.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.9 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 13.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 27.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 1.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 1.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.4 | 1.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 2.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 2.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 1.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 2.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 3.1 | GO:0005343 | sodium:amino acid symporter activity(GO:0005283) organic acid:sodium symporter activity(GO:0005343) |
| 0.3 | 1.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 3.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 2.1 | GO:0051429 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 4.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 2.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 0.7 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 2.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 8.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.4 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 0.6 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.9 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) receptor antagonist activity(GO:0048019) |
| 0.1 | 5.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 6.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.6 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.8 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 3.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 2.6 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.0 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.8 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 10.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 3.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 4.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 2.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 1.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 3.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 0.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |