Project
DANIO-CODE
Navigation
Downloads
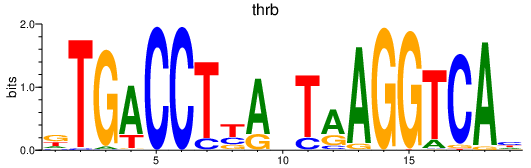
Results for thrb
Z-value: 0.29
Transcription factors associated with thrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
thrb
|
ENSDARG00000021163 | thyroid hormone receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| thrb | dr10_dc_chr19_-_18199573_18199635 | 0.53 | 3.4e-02 | Click! |
Activity profile of thrb motif
Sorted Z-values of thrb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of thrb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_19514967 | 0.41 |
ENSDART00000166073
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr2_+_19515120 | 0.37 |
ENSDART00000166073
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr2_+_19514886 | 0.36 |
ENSDART00000166073
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr13_-_36440277 | 0.35 |
ENSDART00000030133
|
synj2bp
|
synaptojanin 2 binding protein |
| chr15_-_35020312 | 0.35 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr15_-_34550732 | 0.30 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr19_+_30266913 | 0.27 |
ENSDART00000109729
|
si:ch73-130a3.4
|
si:ch73-130a3.4 |
| chr15_-_35020250 | 0.25 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr16_+_17857553 | 0.24 |
ENSDART00000148878
|
them4
|
thioesterase superfamily member 4 |
| chr20_+_27398061 | 0.23 |
ENSDART00000153440
|
BX255913.1
|
ENSDARG00000096709 |
| chr17_+_30826890 | 0.22 |
ENSDART00000149600
|
tpp1
|
tripeptidyl peptidase I |
| chr6_+_60117223 | 0.22 |
ENSDART00000008243
|
prelid3b
|
PRELI domain containing 3 |
| chr2_-_5287507 | 0.21 |
ENSDART00000063384
|
phb2a
|
prohibitin 2a |
| chr3_+_34716635 | 0.19 |
ENSDART00000055263
|
itga3a
|
integrin, alpha 3a |
| chr2_-_5287404 | 0.18 |
ENSDART00000063384
|
phb2a
|
prohibitin 2a |
| chr14_+_37506462 | 0.14 |
ENSDART00000174790
ENSDART00000179610 ENSDART00000179048 |
CU104743.1
|
ENSDARG00000109062 |
| chr6_-_42292607 | 0.02 |
ENSDART00000064969
|
tex264a
|
testis expressed 264a |
| chr24_-_1026977 | 0.01 |
ENSDART00000147508
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 1.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 1.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |