Project
DANIO-CODE
Navigation
Downloads
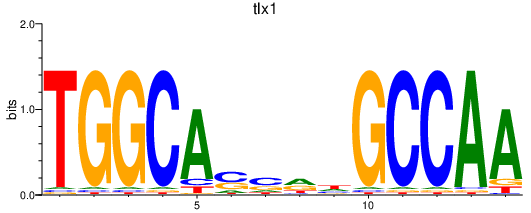
Results for tlx1
Z-value: 1.56
Transcription factors associated with tlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tlx1
|
ENSDARG00000003965 | T cell leukemia homeobox 1 |
Activity profile of tlx1 motif
Sorted Z-values of tlx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tlx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_56970554 | 7.44 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr5_+_12786897 | 5.91 |
|
|
|
| chr19_-_30816780 | 4.74 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr2_+_55859099 | 4.26 |
ENSDART00000097753
ENSDART00000141688 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr16_+_23482744 | 4.06 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr2_-_8219329 | 3.96 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr21_-_34623741 | 3.80 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr16_+_16941228 | 3.78 |
ENSDART00000142155
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr15_-_35034668 | 3.70 |
ENSDART00000153787
|
rnf183
|
ring finger protein 183 |
| chr17_+_52736192 | 3.67 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr5_-_56984800 | 3.67 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr7_-_24201833 | 3.44 |
ENSDART00000121684
|
nat8l
|
N-acetyltransferase 8-like |
| chr2_+_49358871 | 3.42 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr19_+_9085377 | 3.22 |
ENSDART00000091443
|
ENSDARG00000062900
|
ENSDARG00000062900 |
| chr18_-_39491932 | 3.05 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr9_-_24431684 | 3.04 |
ENSDART00000039399
|
cavin2a
|
caveolae associated protein 2a |
| chr7_+_56807833 | 2.89 |
ENSDART00000055956
|
enosf1
|
enolase superfamily member 1 |
| chr6_-_49674729 | 2.66 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr20_+_34417665 | 2.64 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr23_-_9556986 | 2.63 |
ENSDART00000139688
|
soga1
|
suppressor of glucose, autophagy associated 1 |
| chr1_+_9612906 | 2.56 |
ENSDART00000152562
|
eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr22_+_38276148 | 2.55 |
ENSDART00000104504
|
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr6_-_39311711 | 2.46 |
|
|
|
| chr11_-_17620732 | 2.44 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr3_-_56969897 | 2.41 |
|
|
|
| chr7_-_34854886 | 2.35 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr17_+_38314814 | 2.31 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr23_+_7085389 | 2.24 |
|
|
|
| chr22_-_28840467 | 2.14 |
|
|
|
| chr6_+_53349584 | 2.09 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr4_-_16417703 | 2.06 |
ENSDART00000013085
|
dcn
|
decorin |
| chr6_+_2849460 | 2.05 |
|
|
|
| chr8_-_31044627 | 2.04 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr10_-_17274395 | 2.02 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr2_-_21960508 | 1.89 |
ENSDART00000027587
|
ralab
|
v-ral simian leukemia viral oncogene homolog Ab (ras related) |
| chr19_-_22182031 | 1.87 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr11_-_22200590 | 1.80 |
ENSDART00000006580
|
tfeb
|
transcription factor EB |
| chr10_+_36710393 | 1.75 |
|
|
|
| chr19_+_43524904 | 1.75 |
|
|
|
| chr12_+_3189489 | 1.74 |
ENSDART00000010569
|
g6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr22_-_11408859 | 1.71 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr9_+_36051713 | 1.70 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr17_-_42108276 | 1.58 |
ENSDART00000110871
ENSDART00000155484 |
nkx2.4a
|
NK2 homeobox 4a |
| chr20_-_15181042 | 1.57 |
ENSDART00000175585
|
prrc2c
|
proline-rich coiled-coil 2C |
| chr24_+_14093524 | 1.57 |
|
|
|
| chr7_+_38008155 | 1.56 |
ENSDART00000169927
|
lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr5_-_45219030 | 1.53 |
|
|
|
| chr17_+_52736535 | 1.48 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr7_+_37105966 | 1.48 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr24_+_26922969 | 1.46 |
|
|
|
| chr1_+_48923897 | 1.46 |
ENSDART00000101004
ENSDART00000164000 ENSDART00000142957 |
col17a1a
|
collagen, type XVII, alpha 1a |
| chr6_-_39311594 | 1.44 |
|
|
|
| chr9_-_23547539 | 1.44 |
ENSDART00000046423
|
arl4cb
|
ADP-ribosylation factor-like 4Cb |
| chr2_-_23431625 | 1.43 |
ENSDART00000157318
|
BX548024.1
|
ENSDARG00000097593 |
| chr8_-_34077387 | 1.40 |
ENSDART00000159208
ENSDART00000040126 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| KN150670v1_-_61400 | 1.39 |
|
|
|
| chr8_+_41639900 | 1.36 |
ENSDART00000136492
|
si:ch211-158d24.4
|
si:ch211-158d24.4 |
| chr8_-_34077790 | 1.34 |
|
|
|
| chr6_-_35417797 | 1.26 |
ENSDART00000016586
|
rgs5a
|
regulator of G protein signaling 5a |
| chr12_+_34615991 | 1.25 |
|
|
|
| chr8_+_43334181 | 1.23 |
ENSDART00000038566
|
fam101a
|
family with sequence similarity 101, member A |
| chr5_-_56856570 | 1.21 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr14_-_1081393 | 1.13 |
|
|
|
| chr17_+_52736844 | 1.12 |
ENSDART00000160507
|
meis2a
|
Meis homeobox 2a |
| chr8_-_3974357 | 1.11 |
ENSDART00000163754
ENSDART00000169474 |
mtmr3
|
myotubularin related protein 3 |
| chr10_+_7678087 | 1.10 |
ENSDART00000160673
|
hint1
|
histidine triad nucleotide binding protein 1 |
| chr22_-_34619727 | 1.10 |
|
|
|
| chr12_+_35911999 | 1.08 |
ENSDART00000167873
|
baiap2b
|
BAI1-associated protein 2b |
| chr9_+_3578084 | 1.03 |
|
|
|
| chr12_+_27035744 | 1.03 |
ENSDART00000025966
|
hoxb6b
|
homeobox B6b |
| chr2_-_30005139 | 0.97 |
ENSDART00000146968
|
cnpy1
|
canopy1 |
| chr25_-_35022528 | 0.95 |
|
|
|
| chr22_-_22391686 | 0.92 |
ENSDART00000158555
|
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr4_-_72584439 | 0.92 |
ENSDART00000174112
|
si:cabz01071911.4
|
si:cabz01071911.4 |
| chr25_+_4534280 | 0.91 |
ENSDART00000130299
|
deaf1
|
DEAF1 transcription factor |
| chr10_+_8132413 | 0.78 |
ENSDART00000143848
ENSDART00000075485 |
sub1a
|
SUB1 homolog, transcriptional regulator a |
| chr7_-_41188699 | 0.77 |
ENSDART00000150146
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr2_+_45447068 | 0.76 |
ENSDART00000083957
ENSDART00000160867 |
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr10_-_42155708 | 0.73 |
|
|
|
| chr3_+_24352261 | 0.73 |
ENSDART00000059179
|
nptxra
|
neuronal pentraxin receptor a |
| chr3_+_19186494 | 0.72 |
ENSDART00000111528
|
kri1
|
KRI1 homolog |
| chr14_-_32163041 | 0.72 |
ENSDART00000034883
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr14_+_31525014 | 0.68 |
ENSDART00000039880
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr1_-_22066408 | 0.68 |
ENSDART00000134719
ENSDART00000166891 |
prom1b
|
prominin 1 b |
| chr5_-_45219194 | 0.66 |
|
|
|
| chr4_-_4241386 | 0.64 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr5_+_424486 | 0.62 |
ENSDART00000170350
|
thap1
|
THAP domain containing, apoptosis associated protein 1 |
| chr14_-_28126346 | 0.62 |
ENSDART00000054062
|
nek12
|
NIMA-related kinase 12 |
| chr18_-_16933517 | 0.59 |
|
|
|
| chr1_+_34810895 | 0.58 |
|
|
|
| chr1_-_27826175 | 0.58 |
ENSDART00000121758
|
ednrba
|
endothelin receptor Ba |
| chr25_-_31049247 | 0.57 |
ENSDART00000009126
|
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr6_-_19812881 | 0.56 |
ENSDART00000151152
|
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr2_+_17116697 | 0.54 |
|
|
|
| chr17_-_22047445 | 0.53 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr3_-_34470137 | 0.47 |
ENSDART00000055259
|
nmrk1
|
nicotinamide riboside kinase 1 |
| chr23_-_38565866 | 0.46 |
|
|
|
| chr17_-_15594907 | 0.44 |
|
|
|
| chr8_+_21974863 | 0.43 |
|
|
|
| chr19_+_43524684 | 0.39 |
|
|
|
| chr6_+_23462412 | 0.39 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr5_-_56856527 | 0.36 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr18_+_41570495 | 0.31 |
ENSDART00000169621
ENSDART00000162052 |
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr25_-_34635697 | 0.31 |
ENSDART00000113870
|
ENSDARG00000075379
|
ENSDARG00000075379 |
| chr23_-_18646217 | 0.31 |
ENSDART00000138599
|
ssr4
|
signal sequence receptor, delta |
| chr8_-_31044429 | 0.28 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr14_-_2342593 | 0.25 |
|
|
|
| chr9_+_36051871 | 0.22 |
ENSDART00000144141
|
rcan1a
|
regulator of calcineurin 1a |
| chr5_+_30683387 | 0.21 |
ENSDART00000133743
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr6_-_39201507 | 0.20 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr3_+_61158991 | 0.19 |
ENSDART00000126417
|
bri3
|
brain protein I3 |
| chr7_-_73622585 | 0.15 |
ENSDART00000126541
|
zgc:173552
|
zgc:173552 |
| chr3_-_41369324 | 0.13 |
|
|
|
| chr8_+_9660683 | 0.08 |
ENSDART00000091848
|
gripap1
|
GRIP1 associated protein 1 |
| chr6_+_54857805 | 0.07 |
|
|
|
| chr18_+_7584079 | 0.07 |
ENSDART00000163709
|
zgc:77650
|
zgc:77650 |
| chr10_+_572931 | 0.07 |
ENSDART00000129856
ENSDART00000110384 |
smad4a
|
SMAD family member 4a |
| chr6_+_33261144 | 0.04 |
ENSDART00000157428
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.7 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.7 | 4.0 | GO:0051963 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.5 | 2.4 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.4 | 1.1 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.3 | 1.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 1.7 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.2 | 1.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.1 | GO:2000251 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.7 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 1.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.8 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 2.1 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.1 | 2.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 1.7 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 3.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 6.3 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.1 | 0.8 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.8 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 3.4 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 0.6 | GO:0042310 | vasoconstriction(GO:0042310) |
| 0.1 | 1.1 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.0 | 2.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.7 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 2.7 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 5.1 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 2.9 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 1.0 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.6 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 3.8 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 2.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.9 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.0 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.5 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 3.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 0.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 1.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 3.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 3.0 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 3.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.4 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 4.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 19.2 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 4.2 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.6 | 2.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.4 | 1.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 1.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 4.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 4.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 3.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 2.6 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.4 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 1.7 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.1 | 2.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 3.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 2.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 7.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 3.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 1.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 4.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 3.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 7.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 3.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 0.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |