Project
DANIO-CODE
Navigation
Downloads
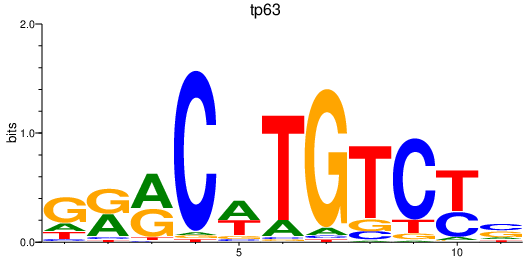
Results for tp63
Z-value: 1.04
Transcription factors associated with tp63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tp63
|
ENSDARG00000044356 | tumor protein p63 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tp63 | dr10_dc_chr6_+_28886741_28886783 | 0.93 | 2.7e-07 | Click! |
Activity profile of tp63 motif
Sorted Z-values of tp63 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tp63
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_25157675 | 5.05 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr24_+_35676505 | 3.21 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr3_+_36146072 | 3.10 |
ENSDART00000148444
|
zgc:86896
|
zgc:86896 |
| chr9_+_41720118 | 2.05 |
|
|
|
| chr8_+_30491476 | 2.04 |
ENSDART00000062073
|
ENSDARG00000042329
|
ENSDARG00000042329 |
| chr9_+_41301317 | 2.01 |
ENSDART00000000250
|
slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr23_+_14058972 | 1.90 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr11_-_80746 | 1.88 |
|
|
|
| chr17_-_14663139 | 1.80 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr19_+_22478256 | 1.80 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| KN150108v1_-_8893 | 1.78 |
ENSDART00000167033
|
ENSDARG00000100513
|
ENSDARG00000100513 |
| chr7_+_69418011 | 1.76 |
ENSDART00000166105
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr16_+_21098948 | 1.72 |
ENSDART00000006043
|
hoxa11b
|
homeobox A11b |
| chr10_-_5135388 | 1.68 |
ENSDART00000108587
|
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr1_+_46794847 | 1.59 |
ENSDART00000140846
|
lrrc58a
|
leucine rich repeat containing 58a |
| chr13_-_33137075 | 1.55 |
ENSDART00000138096
|
oip5-as1
|
oip5 antisense RNA 1 |
| chr20_-_42821812 | 1.41 |
ENSDART00000138957
|
gpr31
|
G protein-coupled receptor 31 |
| chr20_-_1293519 | 1.37 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr3_+_42380497 | 1.34 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr9_-_15453448 | 1.34 |
ENSDART00000124346
ENSDART00000010521 ENSDART00000103753 |
fn1a
|
fibronectin 1a |
| chr5_+_23585492 | 1.29 |
ENSDART00000135083
|
tp53
|
tumor protein p53 |
| chr14_-_9216303 | 1.17 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr15_-_10532992 | 1.17 |
ENSDART00000175825
|
tenm4
|
teneurin transmembrane protein 4 |
| chr8_-_4638117 | 1.10 |
ENSDART00000019828
|
zgc:63587
|
zgc:63587 |
| chr17_-_43475725 | 1.09 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr21_+_45693978 | 1.03 |
ENSDART00000160324
|
CABZ01102047.1
|
ENSDARG00000099648 |
| chr23_+_25224499 | 0.96 |
|
|
|
| chr22_+_16279764 | 0.96 |
|
|
|
| chr11_-_11534819 | 0.90 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr16_+_21099550 | 0.85 |
|
|
|
| chr19_-_31448000 | 0.84 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr18_+_17622557 | 0.77 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr4_+_23406078 | 0.77 |
ENSDART00000139543
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr8_-_12253026 | 0.76 |
ENSDART00000091612
|
dab2ipa
|
DAB2 interacting protein a |
| chr12_+_22455344 | 0.72 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr20_-_25537241 | 0.68 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr10_+_5135537 | 0.67 |
ENSDART00000132627
ENSDART00000082295 |
zgc:113274
|
zgc:113274 |
| chr3_+_21928819 | 0.65 |
ENSDART00000154622
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr9_+_50609926 | 0.65 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr16_+_41767229 | 0.58 |
ENSDART00000075937
|
mtdhb
|
metadherin b |
| chr10_-_40905193 | 0.58 |
ENSDART00000172089
|
pcna
|
proliferating cell nuclear antigen |
| chr24_+_41009372 | 0.42 |
ENSDART00000165952
|
CU633479.3
|
ENSDARG00000099523 |
| chr16_+_21099213 | 0.41 |
ENSDART00000006043
|
hoxa11b
|
homeobox A11b |
| chr12_+_44829938 | 0.35 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr10_-_40904685 | 0.33 |
|
|
|
| chr18_-_17410180 | 0.26 |
ENSDART00000060949
|
zfpm1
|
zinc finger protein, FOG family member 1 |
| chr25_-_35530474 | 0.25 |
|
|
|
| chr12_+_26579396 | 0.24 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr6_+_3120600 | 0.22 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr10_-_40905363 | 0.21 |
ENSDART00000076304
|
pcna
|
proliferating cell nuclear antigen |
| chr4_-_23405867 | 0.20 |
|
|
|
| chr19_+_48166365 | 0.19 |
|
|
|
| chr25_-_35742590 | 0.17 |
ENSDART00000073399
|
faap24
|
Fanconi anemia core complex associated protein 24 |
| chr12_+_26579286 | 0.16 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr25_+_31501347 | 0.13 |
ENSDART00000153968
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr20_+_22813503 | 0.06 |
|
|
|
| chr3_-_27989221 | 0.04 |
ENSDART00000151143
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.3 | 1.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 1.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 2.0 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.3 | 0.8 | GO:0055091 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) reverse cholesterol transport(GO:0043691) phospholipid homeostasis(GO:0055091) |
| 0.3 | 0.8 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.1 | 0.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.8 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 3.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.3 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 3.4 | GO:0033993 | response to lipid(GO:0033993) |
| 0.0 | 1.8 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 0.1 | GO:1901216 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 1.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 3.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.7 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 2.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.9 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.5 | 5.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 1.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 0.8 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.2 | 1.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.8 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 3.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.8 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 0.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 3.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |