Project
DANIO-CODE
Navigation
Downloads
Results for twist1a+twist1b+twist3
Z-value: 0.45
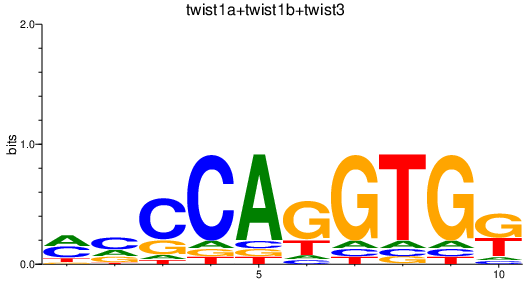
Transcription factors associated with twist1a+twist1b+twist3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
twist3
|
ENSDARG00000019646 | twist3 |
|
twist1a
|
ENSDARG00000030402 | twist family bHLH transcription factor 1a |
|
twist1b
|
ENSDARG00000076010 | twist family bHLH transcription factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| twist1a | dr10_dc_chr19_-_2399429_2399450 | 0.45 | 7.9e-02 | Click! |
| twist1b | dr10_dc_chr16_+_19926776_19926808 | 0.18 | 5.1e-01 | Click! |
| twist3 | dr10_dc_chr23_+_2665_2671 | 0.07 | 7.8e-01 | Click! |
Activity profile of twist1a+twist1b+twist3 motif
Sorted Z-values of twist1a+twist1b+twist3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of twist1a+twist1b+twist3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_17735989 | 1.06 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr9_-_40212864 | 0.72 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr14_-_30747448 | 0.67 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr14_-_30747259 | 0.60 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr14_+_47229400 | 0.57 |
ENSDART00000136045
|
mgst2
|
microsomal glutathione S-transferase 2 |
| chr17_-_25865861 | 0.55 |
|
|
|
| chr20_-_22036656 | 0.46 |
ENSDART00000152290
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr13_-_31165867 | 0.43 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr4_-_12792010 | 0.41 |
ENSDART00000067131
|
irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr1_-_5284684 | 0.41 |
|
|
|
| chr4_-_17033790 | 0.39 |
|
|
|
| chr9_-_40212643 | 0.34 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr20_+_36720896 | 0.33 |
ENSDART00000062895
|
srp9
|
signal recognition particle 9 |
| chr16_-_31835463 | 0.31 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr13_-_47767713 | 0.29 |
ENSDART00000045475
|
itga9
|
integrin, alpha 9 |
| chr15_-_16140187 | 0.28 |
ENSDART00000080413
|
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr6_+_30469823 | 0.23 |
ENSDART00000121492
|
FP236735.1
|
ENSDARG00000087805 |
| chr18_-_40462054 | 0.22 |
|
|
|
| chr14_-_30747372 | 0.22 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr13_-_11667628 | 0.22 |
ENSDART00000079392
|
zmp:0000000662
|
zmp:0000000662 |
| chr6_-_55411751 | 0.19 |
ENSDART00000159246
ENSDART00000158929 |
ctsa
|
cathepsin A |
| chr3_-_22082170 | 0.19 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr20_-_22293402 | 0.17 |
ENSDART00000047624
|
tmem165
|
transmembrane protein 165 |
| chr13_+_25356121 | 0.15 |
ENSDART00000057689
|
bag3
|
BCL2-associated athanogene 3 |
| chr16_-_36061564 | 0.14 |
ENSDART00000175847
|
CABZ01065723.1
|
ENSDARG00000105999 |
| chr9_-_40213104 | 0.14 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr13_+_25356184 | 0.12 |
ENSDART00000057689
|
bag3
|
BCL2-associated athanogene 3 |
| chr6_-_10492439 | 0.12 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr3_+_24059652 | 0.10 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr1_-_43323814 | 0.10 |
ENSDART00000148932
|
si:ch73-109d9.1
|
si:ch73-109d9.1 |
| chr7_+_20829815 | 0.09 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr21_-_1412006 | 0.08 |
|
|
|
| chr21_-_26678542 | 0.07 |
ENSDART00000053794
|
banf1
|
barrier to autointegration factor 1 |
| chr4_+_1680973 | 0.05 |
ENSDART00000149826
ENSDART00000166437 ENSDART00000031135 ENSDART00000172300 ENSDART00000067446 |
slc38a4
|
solute carrier family 38, member 4 |
| chr19_+_26838851 | 0.04 |
ENSDART00000136244
|
npr1a
|
natriuretic peptide receptor 1a |
| chr4_+_26507297 | 0.04 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr9_-_40213039 | 0.03 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.4 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.6 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.3 | GO:0010657 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.4 | GO:0014003 | oligodendrocyte development(GO:0014003) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.3 | 1.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 0.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.3 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |