Project
DANIO-CODE
Navigation
Downloads
Results for vax2
Z-value: 0.55
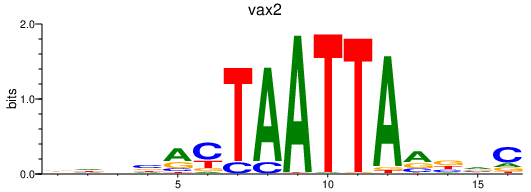
Transcription factors associated with vax2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
vax2
|
ENSDARG00000058702 | ventral anterior homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vax2 | dr10_dc_chr7_-_8635989_8636026 | -0.91 | 1.4e-06 | Click! |
Activity profile of vax2 motif
Sorted Z-values of vax2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of vax2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_34058331 | 2.77 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr5_+_37303599 | 2.57 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr11_-_44539778 | 2.19 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr18_-_40718244 | 1.80 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr11_-_6442588 | 1.55 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr24_+_19270877 | 1.21 |
|
|
|
| chr10_-_4641624 | 1.19 |
ENSDART00000163951
|
plppr1
|
phospholipid phosphatase related 1 |
| chr2_+_6341404 | 1.17 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr12_-_33256754 | 1.15 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_-_33256671 | 1.09 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr2_-_26941084 | 1.07 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr2_-_26941232 | 0.97 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr1_-_54570813 | 0.96 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr4_+_4825461 | 0.95 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr25_-_28630138 | 0.95 |
|
|
|
| chr11_-_18090243 | 0.93 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr12_-_33257026 | 0.93 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_+_33257120 | 0.89 |
|
|
|
| chr2_-_601945 | 0.87 |
|
|
|
| chr12_-_33256599 | 0.87 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr25_-_36678422 | 0.84 |
ENSDART00000087247
|
glg1a
|
golgi glycoprotein 1a |
| chr9_+_23813071 | 0.84 |
|
|
|
| chr3_-_19890717 | 0.81 |
ENSDART00000104118
ENSDART00000170199 |
atxn7l3
|
ataxin 7-like 3 |
| chr25_+_19302983 | 0.78 |
|
|
|
| chr14_+_46362661 | 0.77 |
ENSDART00000025749
|
ttc9c
|
tetratricopeptide repeat domain 9C |
| chr12_-_33256934 | 0.74 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr21_-_13026036 | 0.74 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr15_+_21327206 | 0.71 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr4_-_62022037 | 0.70 |
|
|
|
| chr2_-_26940965 | 0.70 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr20_+_29306863 | 0.70 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr21_+_31113731 | 0.69 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr11_-_38856849 | 0.68 |
ENSDART00000113185
|
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr13_-_36672424 | 0.64 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr20_-_28898117 | 0.61 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr8_+_24276275 | 0.61 |
|
|
|
| chr25_-_13394261 | 0.61 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr10_-_35377548 | 0.59 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr16_-_31386496 | 0.59 |
|
|
|
| chr5_+_37303707 | 0.58 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr19_+_48731233 | 0.56 |
|
|
|
| chr8_+_3364453 | 0.56 |
ENSDART00000164426
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr20_+_29306677 | 0.52 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr13_-_8360779 | 0.51 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr11_-_44539726 | 0.49 |
ENSDART00000173360
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr21_-_2350090 | 0.47 |
ENSDART00000168946
|
si:ch211-241b2.4
|
si:ch211-241b2.4 |
| chr20_+_35176069 | 0.47 |
|
|
|
| chr20_-_37910887 | 0.46 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr19_+_48731140 | 0.45 |
|
|
|
| chr19_+_5562075 | 0.45 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr16_+_23884575 | 0.44 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr15_-_43402935 | 0.44 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr20_-_46079529 | 0.42 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr23_-_25853364 | 0.41 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr4_+_4825628 | 0.40 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr14_+_29601073 | 0.39 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr8_+_26040518 | 0.39 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr2_+_10209233 | 0.39 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr2_+_54267946 | 0.39 |
ENSDART00000015535
|
ctnnbl1
|
catenin, beta like 1 |
| chr11_-_6442490 | 0.39 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr24_+_16402587 | 0.38 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr20_-_38851375 | 0.37 |
|
|
|
| chr21_-_13026077 | 0.35 |
ENSDART00000024616
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr3_-_26657213 | 0.35 |
|
|
|
| chr11_+_5522377 | 0.34 |
ENSDART00000013203
|
cse1l
|
CSE1 chromosome segregation 1-like (yeast) |
| chr23_-_33692244 | 0.32 |
|
|
|
| chr17_+_1080921 | 0.31 |
|
|
|
| chr12_-_49026424 | 0.31 |
|
|
|
| chr10_-_2761480 | 0.30 |
ENSDART00000131749
|
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr10_-_35377482 | 0.30 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr12_+_19159391 | 0.28 |
ENSDART00000153428
|
tnrc6b
|
trinucleotide repeat containing 6b |
| chr17_+_28689850 | 0.27 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr12_+_19159234 | 0.27 |
ENSDART00000153428
|
tnrc6b
|
trinucleotide repeat containing 6b |
| chr2_+_54268010 | 0.26 |
ENSDART00000170799
|
ctnnbl1
|
catenin, beta like 1 |
| chr25_-_12712431 | 0.25 |
ENSDART00000162750
|
ca5a
|
carbonic anhydrase Va |
| chr17_+_1799209 | 0.25 |
ENSDART00000112183
|
cep170b
|
centrosomal protein 170B |
| chr10_-_4641512 | 0.24 |
ENSDART00000163951
|
plppr1
|
phospholipid phosphatase related 1 |
| chr16_-_42105733 | 0.24 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr20_+_29306945 | 0.24 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr17_+_8642373 | 0.23 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr21_+_10757086 | 0.23 |
|
|
|
| chr16_-_42105636 | 0.23 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr14_+_29601252 | 0.23 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr4_+_952596 | 0.23 |
ENSDART00000122535
|
rpap3
|
RNA polymerase II associated protein 3 |
| chr9_-_11705194 | 0.21 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr21_+_17509116 | 0.20 |
|
|
|
| chr3_-_19890791 | 0.20 |
ENSDART00000104118
ENSDART00000170199 |
atxn7l3
|
ataxin 7-like 3 |
| chr1_-_5387933 | 0.20 |
ENSDART00000138891
ENSDART00000176913 |
pard3ba
|
par-3 family cell polarity regulator beta a |
| chr17_+_8642686 | 0.20 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr19_+_21783204 | 0.18 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr9_+_26218474 | 0.17 |
ENSDART00000177394
|
CR391949.1
|
ENSDARG00000107108 |
| chr13_-_36672285 | 0.17 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr25_-_24440292 | 0.17 |
ENSDART00000156805
|
CR854832.1
|
ENSDARG00000096817 |
| chr14_-_14353487 | 0.17 |
ENSDART00000172241
|
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr23_-_25853331 | 0.16 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr3_+_35277555 | 0.16 |
|
|
|
| chr8_-_25015215 | 0.15 |
ENSDART00000170511
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr23_+_28882409 | 0.15 |
ENSDART00000078171
|
pex14
|
peroxisomal biogenesis factor 14 |
| chr6_+_3313515 | 0.14 |
ENSDART00000011785
|
rad54l
|
RAD54-like (S. cerevisiae) |
| chr4_-_1952230 | 0.14 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr11_+_44414503 | 0.14 |
|
|
|
| chr21_-_13875945 | 0.13 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr13_-_36672606 | 0.11 |
ENSDART00000179242
|
sav1
|
salvador family WW domain containing protein 1 |
| chr16_-_15497269 | 0.11 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr2_+_50873843 | 0.09 |
|
|
|
| chr15_-_36490729 | 0.08 |
|
|
|
| chr25_-_12712565 | 0.08 |
ENSDART00000162750
|
ca5a
|
carbonic anhydrase Va |
| chr23_-_2957203 | 0.07 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr20_+_29307283 | 0.07 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr15_-_11799447 | 0.03 |
ENSDART00000161445
|
fkrp
|
fukutin related protein |
| chr10_+_40362746 | 0.02 |
ENSDART00000062795
ENSDART00000113582 |
git2b
|
G protein-coupled receptor kinase interacting ArfGAP 2b |
| chr16_+_40396568 | 0.02 |
ENSDART00000139272
|
BX072576.2
|
ENSDARG00000090181 |
| chr13_+_516546 | 0.01 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr22_+_2074020 | 0.01 |
ENSDART00000106540
|
znf1161
|
zinc finger protein 1161 |
| chr10_-_9294185 | 0.00 |
|
|
|
| chr8_-_26210618 | 0.00 |
ENSDART00000133466
|
CR847503.1
|
ENSDARG00000092715 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 1.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.6 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 3.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 0.5 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 1.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.3 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.0 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 0.4 | GO:0046037 | GMP biosynthetic process(GO:0006177) GTP biosynthetic process(GO:0006183) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0046379 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.4 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 1.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:1901910 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.6 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.1 | 1.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 1.0 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.7 | 4.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 0.9 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 0.6 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 1.2 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 3.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 1.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 1.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |