Project
DANIO-CODE
Navigation
Downloads
Results for vdrb_nr1i2
Z-value: 0.72
Transcription factors associated with vdrb_nr1i2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
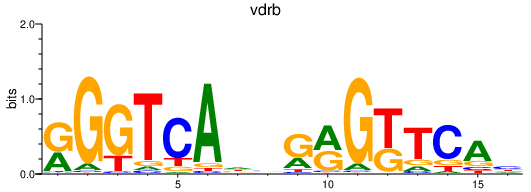
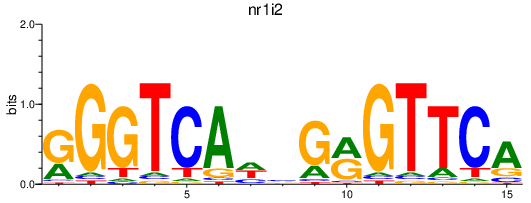
vdrb
|
ENSDARG00000070721 | vitamin D receptor b |
|
nr1i2
|
ENSDARG00000029766 | nuclear receptor subfamily 1, group I, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vdrb | dr10_dc_chr6_-_39008833_39009010 | 0.59 | 1.7e-02 | Click! |
| nr1i2 | dr10_dc_chr9_+_9524547_9524594 | 0.32 | 2.3e-01 | Click! |
Activity profile of vdrb_nr1i2 motif
Sorted Z-values of vdrb_nr1i2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of vdrb_nr1i2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_56692366 | 1.83 |
|
|
|
| chr9_+_10721552 | 1.81 |
ENSDART00000061499
|
cxcr4b
|
chemokine (C-X-C motif), receptor 4b |
| chr6_+_50452131 | 1.48 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr3_-_29846530 | 1.48 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr5_-_60671545 | 1.42 |
|
|
|
| chr13_-_3801122 | 1.34 |
|
|
|
| chr25_+_17774363 | 1.27 |
ENSDART00000136797
|
btbd10a
|
BTB (POZ) domain containing 10a |
| chr2_-_53772219 | 1.25 |
ENSDART00000037557
|
admp
|
anti-dorsalizing morphogenic protein |
| chr7_-_23965038 | 0.97 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr5_-_36349284 | 0.85 |
ENSDART00000047269
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr19_-_43468872 | 0.81 |
|
|
|
| chr21_+_30434147 | 0.80 |
ENSDART00000147375
|
snx12
|
sorting nexin 12 |
| chr7_-_11812634 | 0.80 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr19_+_30210743 | 0.76 |
ENSDART00000167803
|
marcksl1b
|
MARCKS-like 1b |
| chr4_-_13922285 | 0.69 |
ENSDART00000080334
|
yaf2
|
YY1 associated factor 2 |
| chr1_-_18695214 | 0.58 |
|
|
|
| chr25_+_12397532 | 0.57 |
ENSDART00000163508
|
map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr10_-_20524586 | 0.55 |
|
|
|
| chr16_+_19730958 | 0.48 |
ENSDART00000139357
|
sp8b
|
sp8 transcription factor b |
| chr3_+_19186494 | 0.45 |
ENSDART00000111528
|
kri1
|
KRI1 homolog |
| chr6_+_50452229 | 0.43 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr9_-_56692281 | 0.43 |
|
|
|
| chr13_-_1177857 | 0.42 |
ENSDART00000111052
|
olig3
|
oligodendrocyte transcription factor 3 |
| chr23_-_45011360 | 0.42 |
ENSDART00000129138
|
mogat3a
|
monoacylglycerol O-acyltransferase 3a |
| chr19_+_43468063 | 0.40 |
ENSDART00000168263
|
pum1
|
pumilio RNA-binding family member 1 |
| chr21_+_5483017 | 0.39 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr15_-_18110169 | 0.39 |
|
|
|
| chr23_-_18958008 | 0.37 |
ENSDART00000133419
|
CR847953.1
|
ENSDARG00000057403 |
| chr20_+_49273889 | 0.37 |
ENSDART00000166051
ENSDART00000112689 |
crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr7_+_45731077 | 0.36 |
ENSDART00000159700
|
si:ch211-260e23.9
|
si:ch211-260e23.9 |
| chr3_+_19186724 | 0.33 |
ENSDART00000110548
|
kri1
|
KRI1 homolog |
| chr9_-_39190534 | 0.33 |
|
|
|
| chr23_-_45011264 | 0.33 |
ENSDART00000129138
|
mogat3a
|
monoacylglycerol O-acyltransferase 3a |
| chr16_+_19730820 | 0.33 |
ENSDART00000079201
|
sp8b
|
sp8 transcription factor b |
| chr23_+_41349364 | 0.31 |
|
|
|
| chr16_+_32097505 | 0.31 |
ENSDART00000039109
|
leng1
|
leukocyte receptor cluster (LRC) member 1 |
| chr19_+_43468926 | 0.29 |
|
|
|
| chr8_-_3287944 | 0.28 |
ENSDART00000057874
|
FUT9 (1 of many)
|
fucosyltransferase 9 |
| chr3_+_22386709 | 0.27 |
|
|
|
| chr16_+_33041823 | 0.27 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr15_+_2458093 | 0.27 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr22_-_16009772 | 0.26 |
|
|
|
| chr23_+_13106555 | 0.26 |
|
|
|
| chr16_+_19730756 | 0.26 |
ENSDART00000112894
|
sp8b
|
sp8 transcription factor b |
| chr19_-_43468824 | 0.26 |
|
|
|
| chr12_+_14176424 | 0.26 |
ENSDART00000078052
|
mto1
|
mitochondrial tRNA translation optimization 1 |
| chr23_+_42488817 | 0.25 |
ENSDART00000171459
|
cyp2aa2
|
y cytochrome P450, family 2, subfamily AA, polypeptide 2 |
| KN150670v1_-_32201 | 0.24 |
|
|
|
| chr23_-_21490386 | 0.23 |
|
|
|
| chr6_+_50452529 | 0.23 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr14_+_21779325 | 0.22 |
ENSDART00000106147
|
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr15_-_33782909 | 0.22 |
ENSDART00000167705
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr12_+_10096495 | 0.22 |
ENSDART00000152212
ENSDART00000127343 |
smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr6_-_43679553 | 0.22 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr5_+_68943914 | 0.21 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr23_-_3809640 | 0.21 |
|
|
|
| chr19_+_30210491 | 0.19 |
ENSDART00000167803
|
marcksl1b
|
MARCKS-like 1b |
| chr20_+_230394 | 0.19 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr15_+_2457956 | 0.18 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr14_+_21999898 | 0.18 |
|
|
|
| chr6_-_39083081 | 0.17 |
ENSDART00000021520
|
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr21_+_17509116 | 0.17 |
|
|
|
| chr21_-_25719352 | 0.16 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr10_-_14530112 | 0.16 |
ENSDART00000176126
|
galt
|
galactose-1-phosphate uridylyltransferase |
| chr19_+_4102502 | 0.16 |
ENSDART00000168112
|
btr23
|
bloodthirsty-related gene family, member 23 |
| chr23_-_3809591 | 0.15 |
|
|
|
| chr22_+_24362712 | 0.15 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr9_+_49054601 | 0.15 |
ENSDART00000047401
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr16_+_6002342 | 0.15 |
ENSDART00000060519
|
ulk4
|
unc-51 like kinase 4 |
| chr7_+_34235330 | 0.14 |
ENSDART00000173957
|
rfx7
|
regulatory factor X, 7 |
| chr14_+_45774736 | 0.14 |
ENSDART00000139691
|
rnf44
|
ring finger protein 44 |
| chr3_+_15245529 | 0.14 |
|
|
|
| KN150670v1_-_32341 | 0.13 |
|
|
|
| chr5_+_64175494 | 0.13 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr10_-_22834248 | 0.13 |
ENSDART00000079469
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr9_+_32379130 | 0.12 |
|
|
|
| chr16_-_35376945 | 0.12 |
ENSDART00000162518
|
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr14_+_31133551 | 0.12 |
ENSDART00000173088
|
ccdc160
|
coiled-coil domain containing 160 |
| chr17_-_25545464 | 0.12 |
ENSDART00000040032
ENSDART00000163447 |
rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr19_-_4079427 | 0.12 |
ENSDART00000170325
|
map7d1b
|
MAP7 domain containing 1b |
| chr23_-_26150495 | 0.12 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr19_+_177005 | 0.12 |
ENSDART00000111580
|
tmem65
|
transmembrane protein 65 |
| chr14_+_46652294 | 0.12 |
|
|
|
| chr8_+_16954190 | 0.11 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr23_-_3813451 | 0.11 |
|
|
|
| chr21_-_9313788 | 0.11 |
|
|
|
| chr21_-_13131290 | 0.11 |
|
|
|
| chr24_+_9073790 | 0.11 |
|
|
|
| chr5_+_69096325 | 0.11 |
ENSDART00000014649
|
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr7_+_23798237 | 0.11 |
ENSDART00000034218
|
acin1b
|
apoptotic chromatin condensation inducer 1b |
| chr21_+_22842181 | 0.10 |
ENSDART00000065562
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr24_+_17201072 | 0.10 |
ENSDART00000024722
|
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr13_+_18376642 | 0.10 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr16_-_7310925 | 0.10 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr7_+_25765060 | 0.10 |
|
|
|
| chr4_+_30012767 | 0.09 |
ENSDART00000150383
|
znf1128
|
zinc finger protein 1128 |
| chr10_+_17113205 | 0.09 |
|
|
|
| chr16_+_19731017 | 0.09 |
ENSDART00000139357
|
sp8b
|
sp8 transcription factor b |
| chr5_-_1131341 | 0.08 |
ENSDART00000168336
|
si:zfos-128g4.1
|
si:zfos-128g4.1 |
| chr15_+_24821561 | 0.08 |
ENSDART00000156424
|
cpda
|
carboxypeptidase D, a |
| chr4_-_16356071 | 0.08 |
ENSDART00000079521
|
kera
|
keratocan |
| chr1_-_24988461 | 0.07 |
ENSDART00000054230
|
fgg
|
fibrinogen gamma chain |
| chr17_-_37107310 | 0.07 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr12_-_41128584 | 0.07 |
|
|
|
| chr8_-_2529834 | 0.07 |
|
|
|
| chr23_-_28881880 | 0.07 |
ENSDART00000078167
|
dffa
|
DNA fragmentation factor, alpha polypeptide |
| chr13_-_1177745 | 0.07 |
ENSDART00000111052
|
olig3
|
oligodendrocyte transcription factor 3 |
| chr16_+_42868905 | 0.06 |
ENSDART00000156601
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr19_-_33301572 | 0.06 |
ENSDART00000052080
|
laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr1_+_23093114 | 0.06 |
|
|
|
| chr3_+_15645543 | 0.06 |
|
|
|
| chr5_+_64175669 | 0.06 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr2_+_393439 | 0.06 |
ENSDART00000122138
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr4_-_74789596 | 0.06 |
ENSDART00000174356
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr6_-_9394499 | 0.06 |
ENSDART00000013066
|
ercc3
|
excision repair cross-complementation group 3 |
| chr19_+_43468369 | 0.06 |
ENSDART00000165202
|
pum1
|
pumilio RNA-binding family member 1 |
| chr5_-_18548689 | 0.06 |
ENSDART00000137802
|
fam214b
|
family with sequence similarity 214, member B |
| chr23_-_3809488 | 0.05 |
|
|
|
| chr24_-_36227334 | 0.05 |
|
|
|
| chr8_-_13603209 | 0.05 |
ENSDART00000080875
|
b3gnt3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
| chr12_+_16845837 | 0.05 |
ENSDART00000138174
|
slc16a12b
|
solute carrier family 16, member 12b |
| chr15_+_2710241 | 0.05 |
ENSDART00000157120
|
CR382327.3
|
ENSDARG00000097494 |
| chr22_+_24362832 | 0.05 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr9_-_6230087 | 0.05 |
ENSDART00000032368
|
tmtops2b
|
teleost multiple tissue opsin 2b |
| chr23_-_28881717 | 0.05 |
ENSDART00000078167
|
dffa
|
DNA fragmentation factor, alpha polypeptide |
| chr12_+_30538113 | 0.04 |
ENSDART00000179355
|
thbs2b
|
thrombospondin 2b |
| chr3_-_31743330 | 0.04 |
ENSDART00000155298
|
limd2
|
LIM domain containing 2 |
| chr24_+_38267919 | 0.04 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr1_-_29885008 | 0.04 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr24_+_19842162 | 0.04 |
ENSDART00000123031
|
CR381686.3
|
ENSDARG00000094073 |
| chr3_-_2686533 | 0.04 |
|
|
|
| chr14_+_44312764 | 0.04 |
ENSDART00000098643
|
tomm5
|
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr5_+_67129480 | 0.03 |
|
|
|
| chr20_+_19093073 | 0.03 |
ENSDART00000147105
|
tdh
|
L-threonine dehydrogenase |
| chr5_+_50432868 | 0.03 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr14_-_20766762 | 0.03 |
ENSDART00000164373
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr2_+_30932612 | 0.03 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr15_+_29460803 | 0.02 |
ENSDART00000155198
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr23_+_25381458 | 0.02 |
ENSDART00000140463
|
esyt1a
|
extended synaptotagmin-like protein 1a |
| chr12_+_16848206 | 0.02 |
|
|
|
| chr14_-_21779536 | 0.02 |
|
|
|
| chr14_+_30413350 | 0.02 |
ENSDART00000009385
|
yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr6_+_41455676 | 0.01 |
ENSDART00000007798
ENSDART00000162135 |
twf2a
|
twinfilin actin-binding protein 2a |
| chr20_+_45530807 | 0.01 |
|
|
|
| chr3_+_15605789 | 0.01 |
ENSDART00000140160
|
BX914211.1
|
ENSDARG00000094116 |
| chr4_-_16356247 | 0.01 |
ENSDART00000079521
|
kera
|
keratocan |
| chr5_-_18548376 | 0.01 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr18_-_44415397 | 0.01 |
|
|
|
| chr14_-_25011874 | 0.01 |
ENSDART00000159569
|
htr4
|
5-hydroxytryptamine receptor 4 |
| chr23_-_3808881 | 0.01 |
|
|
|
| chr3_-_29846789 | 0.00 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr1_-_44513922 | 0.00 |
ENSDART00000014727
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0002686 | negative regulation of leukocyte migration(GO:0002686) |
| 0.2 | 1.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.2 | 1.3 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.1 | 0.8 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 0.3 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 1.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.5 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.2 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 2.1 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.4 | GO:0071007 | Prp19 complex(GO:0000974) U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 1.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.7 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.4 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.2 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.2 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 1.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 1.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |