Project
DANIO-CODE
Navigation
Downloads
Results for ved
Z-value: 0.40
Transcription factors associated with ved
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ved
|
ENSDARG00000102389 | ventrally expressed dharma/bozozok antagonist |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ved | dr10_dc_chr10_-_45155460_45155607 | -0.23 | 4.0e-01 | Click! |
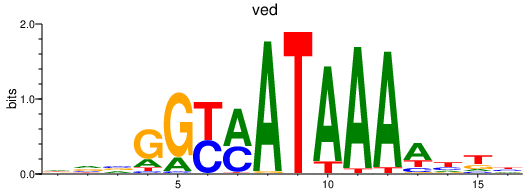
Activity profile of ved motif
Sorted Z-values of ved motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ved
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_32394653 | 0.47 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr7_-_29021031 | 0.35 |
ENSDART00000086753
|
dapk2a
|
death-associated protein kinase 2a |
| chr3_+_44928592 | 0.30 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr25_+_19992389 | 0.28 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr25_+_8936284 | 0.26 |
ENSDART00000154207
|
im:7145024
|
im:7145024 |
| chr7_+_50491026 | 0.26 |
ENSDART00000165037
|
si:ch73-380l10.2
|
si:ch73-380l10.2 |
| chr9_-_34450885 | 0.24 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr5_-_23211957 | 0.23 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr6_-_40700033 | 0.21 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr3_+_44928323 | 0.21 |
ENSDART00000170913
|
zgc:112146
|
zgc:112146 |
| chr25_+_30701751 | 0.19 |
|
|
|
| chr22_-_11463487 | 0.19 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr14_+_34150130 | 0.18 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr21_+_20360180 | 0.18 |
ENSDART00000003299
|
ENSDARG00000025174
|
ENSDARG00000025174 |
| chr3_+_32394550 | 0.17 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr24_-_6129575 | 0.17 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr18_-_26131535 | 0.17 |
|
|
|
| chr19_-_7306136 | 0.17 |
ENSDART00000133179
|
col11a2
|
collagen, type XI, alpha 2 |
| chr5_+_58017820 | 0.16 |
ENSDART00000083015
|
ccdc84
|
coiled-coil domain containing 84 |
| chr11_-_25019899 | 0.16 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr10_-_25448712 | 0.16 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr15_-_23973374 | 0.16 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr8_-_38126693 | 0.16 |
ENSDART00000112331
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr24_-_38758093 | 0.16 |
|
|
|
| chr1_-_8801877 | 0.15 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr21_-_11877525 | 0.15 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr6_+_102073 | 0.15 |
ENSDART00000151209
|
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr3_-_30984999 | 0.15 |
ENSDART00000058955
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr2_+_35750334 | 0.15 |
ENSDART00000052666
|
rasal2
|
RAS protein activator like 2 |
| chr1_-_52833379 | 0.14 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr11_-_16067646 | 0.14 |
|
|
|
| chr23_+_28563324 | 0.14 |
ENSDART00000006657
|
huwe1
|
HECT, UBA and WWE domain containing 1 |
| chr25_-_12538613 | 0.14 |
ENSDART00000162463
|
zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr5_-_12213712 | 0.14 |
ENSDART00000099749
|
CLDN22
|
claudin 22 |
| chr19_-_7306228 | 0.13 |
ENSDART00000104799
|
col11a2
|
collagen, type XI, alpha 2 |
| chr2_-_17564676 | 0.13 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr6_+_27676752 | 0.13 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr12_+_13167781 | 0.13 |
ENSDART00000160664
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr6_+_4379989 | 0.13 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr18_+_36683567 | 0.13 |
ENSDART00000098972
|
capn12
|
calpain 12 |
| chr1_+_40763274 | 0.12 |
ENSDART00000135280
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr24_-_32115473 | 0.12 |
ENSDART00000168419
|
rsu1
|
Ras suppressor protein 1 |
| chr5_-_57053687 | 0.12 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr4_+_16020464 | 0.12 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr5_-_68727127 | 0.12 |
ENSDART00000023983
|
psat1
|
phosphoserine aminotransferase 1 |
| chr15_-_4589565 | 0.12 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr15_+_44069851 | 0.12 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr1_+_29027527 | 0.12 |
ENSDART00000142184
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr14_+_14536088 | 0.11 |
ENSDART00000158291
|
slbp
|
stem-loop binding protein |
| chr2_+_23383492 | 0.11 |
ENSDART00000170669
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr19_-_7439294 | 0.11 |
|
|
|
| chr22_-_28840467 | 0.11 |
|
|
|
| chr2_-_41035670 | 0.11 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr15_+_30442890 | 0.11 |
ENSDART00000048847
|
nos2b
|
nitric oxide synthase 2b, inducible |
| chr4_-_20414881 | 0.11 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr7_-_38363533 | 0.11 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr19_+_1922930 | 0.11 |
ENSDART00000013217
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr6_-_6818607 | 0.11 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr17_-_25612341 | 0.11 |
ENSDART00000126201
ENSDART00000105503 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr7_-_47990610 | 0.11 |
ENSDART00000147968
|
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr10_-_43874597 | 0.11 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr24_+_2678808 | 0.11 |
|
|
|
| chr13_-_44883668 | 0.11 |
|
|
|
| chr16_-_21238288 | 0.11 |
ENSDART00000139737
|
cbx3b
|
chromobox homolog 3b |
| chr2_+_1881022 | 0.11 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr25_+_34791665 | 0.11 |
|
|
|
| chr20_-_51354971 | 0.10 |
|
|
|
| chr6_-_13586850 | 0.10 |
ENSDART00000111102
|
chpfb
|
chondroitin polymerizing factor b |
| chr4_+_4825628 | 0.10 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr21_-_3548863 | 0.10 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr18_-_25784843 | 0.10 |
ENSDART00000103046
|
zgc:162879
|
zgc:162879 |
| chr8_-_25015215 | 0.10 |
ENSDART00000170511
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr22_-_22139268 | 0.10 |
ENSDART00000101659
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr13_+_45294725 | 0.10 |
ENSDART00000145016
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr23_+_43916552 | 0.10 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr14_+_38509671 | 0.10 |
|
|
|
| chr23_-_3815871 | 0.10 |
ENSDART00000137826
|
hmga1a
|
high mobility group AT-hook 1a |
| chr12_+_10015987 | 0.10 |
ENSDART00000078497
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr23_+_22858063 | 0.10 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr13_+_40311601 | 0.10 |
ENSDART00000148510
|
got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr21_-_15833030 | 0.09 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr3_-_29779725 | 0.09 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr10_+_44853207 | 0.09 |
ENSDART00000169466
|
scarb1
|
scavenger receptor class B, member 1 |
| chr21_-_22435721 | 0.09 |
|
|
|
| chr25_+_19992328 | 0.09 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr4_+_4825461 | 0.09 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr13_-_15662599 | 0.09 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2-associated athanogene 5 |
| chr4_+_74816506 | 0.09 |
|
|
|
| chr3_-_29779552 | 0.09 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr22_-_4379068 | 0.09 |
|
|
|
| chr19_-_17400395 | 0.09 |
ENSDART00000171284
|
sf3a3
|
splicing factor 3a, subunit 3 |
| chr21_-_39035904 | 0.09 |
ENSDART00000075935
|
vtnb
|
vitronectin b |
| chr12_+_26579396 | 0.09 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr18_+_9755131 | 0.09 |
|
|
|
| chr10_-_30016761 | 0.09 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr1_+_54231447 | 0.09 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr14_-_2819498 | 0.08 |
ENSDART00000090196
|
slc35a4
|
solute carrier family 35, member A4 |
| chr16_-_43630591 | 0.08 |
|
|
|
| chr6_+_46968199 | 0.08 |
|
|
|
| chr23_-_18760927 | 0.08 |
ENSDART00000051182
|
arhgap4b
|
Rho GTPase activating protein 4b |
| chr3_+_18263447 | 0.08 |
ENSDART00000127796
|
gaa
|
glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) |
| chr3_+_13450981 | 0.08 |
ENSDART00000162124
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr18_-_26131127 | 0.08 |
ENSDART00000163226
ENSDART00000004692 |
idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr18_-_25784699 | 0.08 |
ENSDART00000103046
|
zgc:162879
|
zgc:162879 |
| chr10_-_24401876 | 0.08 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr7_+_23735703 | 0.08 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr1_+_33510660 | 0.08 |
ENSDART00000147201
|
slc5a7a
|
solute carrier family 5 (sodium/choline cotransporter), member 7a |
| chr7_+_1337856 | 0.08 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr3_+_40022244 | 0.08 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr9_+_55963230 | 0.08 |
|
|
|
| chr23_-_43916621 | 0.08 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr20_-_27834646 | 0.08 |
ENSDART00000138139
|
zgc:153157
|
zgc:153157 |
| chr19_+_39100820 | 0.08 |
|
|
|
| chr23_-_27579257 | 0.08 |
ENSDART00000137229
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr17_+_32390603 | 0.07 |
ENSDART00000156051
|
dhx32b
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32b |
| chr11_-_44360453 | 0.07 |
ENSDART00000171837
|
tbce
|
tubulin folding cofactor E |
| chr23_+_22857958 | 0.07 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr10_+_15382647 | 0.07 |
ENSDART00000046274
|
trappc13
|
trafficking protein particle complex 13 |
| chr18_+_17594381 | 0.07 |
ENSDART00000010998
|
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr13_-_35908796 | 0.07 |
ENSDART00000084929
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr19_+_22478256 | 0.07 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr23_+_43916520 | 0.07 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr2_-_36058327 | 0.07 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr11_-_33970180 | 0.07 |
ENSDART00000164353
|
lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr10_-_31619761 | 0.07 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr11_+_6118826 | 0.07 |
ENSDART00000159680
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr13_-_17729510 | 0.07 |
ENSDART00000170583
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| KN149795v1_-_10712 | 0.07 |
|
|
|
| chr3_-_30985152 | 0.07 |
ENSDART00000058955
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr20_+_52740555 | 0.07 |
ENSDART00000110777
|
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr15_+_42329433 | 0.07 |
ENSDART00000130404
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr22_-_22139099 | 0.07 |
|
|
|
| chr1_+_44971899 | 0.07 |
ENSDART00000149565
|
trappc5
|
trafficking protein particle complex 5 |
| chr6_+_30385355 | 0.07 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr3_+_39437613 | 0.07 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr18_+_20058174 | 0.07 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr16_-_43235448 | 0.06 |
ENSDART00000165293
|
slc25a40
|
solute carrier family 25, member 40 |
| chr3_+_24059652 | 0.06 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr16_+_53078999 | 0.06 |
ENSDART00000148435
|
trip13
|
thyroid hormone receptor interactor 13 |
| chr24_-_32110808 | 0.06 |
ENSDART00000159034
|
rsu1
|
Ras suppressor protein 1 |
| chr12_+_13167838 | 0.06 |
ENSDART00000160664
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr15_-_4589523 | 0.06 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr13_+_11303809 | 0.06 |
ENSDART00000169895
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr14_-_4014025 | 0.06 |
ENSDART00000157782
|
irf2
|
interferon regulatory factor 2 |
| chr20_-_40357302 | 0.06 |
|
|
|
| chr8_+_25940883 | 0.06 |
ENSDART00000140626
|
ENSDARG00000061023
|
ENSDARG00000061023 |
| chr9_-_56440750 | 0.06 |
ENSDART00000161247
|
sowahca
|
sosondowah ankyrin repeat domain family Ca |
| chr24_-_32110851 | 0.06 |
ENSDART00000159034
|
rsu1
|
Ras suppressor protein 1 |
| chr25_-_35094522 | 0.06 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr13_-_15662476 | 0.06 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2-associated athanogene 5 |
| chr2_-_28319823 | 0.06 |
ENSDART00000020367
|
zgc:56556
|
zgc:56556 |
| chr6_+_49552893 | 0.06 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr13_-_44883880 | 0.06 |
|
|
|
| chr1_+_40762892 | 0.06 |
ENSDART00000010211
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr23_-_1562603 | 0.06 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr15_-_23973415 | 0.06 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr10_+_27106476 | 0.06 |
ENSDART00000012717
|
ehd1b
|
EH-domain containing 1b |
| chr7_-_70466713 | 0.05 |
|
|
|
| chr4_-_5766814 | 0.05 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr24_-_9857510 | 0.05 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr1_-_51658383 | 0.05 |
ENSDART00000132638
|
ENSDARG00000059948
|
ENSDARG00000059948 |
| chr10_+_39142185 | 0.05 |
|
|
|
| chr14_-_47226179 | 0.05 |
ENSDART00000056734
ENSDART00000143158 |
setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr16_+_42567707 | 0.05 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr1_-_8801818 | 0.05 |
ENSDART00000008682
ENSDART00000157814 |
micall2b
|
mical-like 2b |
| chr13_-_29290894 | 0.05 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr18_+_1778654 | 0.05 |
ENSDART00000176491
|
CABZ01073163.1
|
ENSDARG00000107400 |
| chr23_-_10787670 | 0.05 |
ENSDART00000177571
|
foxp1a
|
forkhead box P1a |
| chr1_-_37639802 | 0.05 |
|
|
|
| chr6_-_6818925 | 0.05 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr2_+_57894141 | 0.05 |
ENSDART00000171264
ENSDART00000163278 |
si:ch211-155e24.3
|
si:ch211-155e24.3 |
| chr5_-_7901538 | 0.04 |
ENSDART00000159718
|
spef2
|
sperm flagellar 2 |
| chr15_-_19192862 | 0.04 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr24_+_39213400 | 0.04 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr18_-_26131468 | 0.04 |
|
|
|
| chr4_+_5246465 | 0.04 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr6_+_10491884 | 0.04 |
ENSDART00000151017
|
BX936397.2
|
ENSDARG00000096373 |
| chr2_+_22753590 | 0.04 |
ENSDART00000171853
|
LMO4
|
zgc:56628 |
| chr23_+_24711233 | 0.04 |
|
|
|
| chr20_-_15029379 | 0.04 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr14_-_30642549 | 0.04 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr23_-_24772871 | 0.04 |
ENSDART00000104031
|
sdf4
|
stromal cell derived factor 4 |
| chr19_-_17400440 | 0.04 |
ENSDART00000160433
|
sf3a3
|
splicing factor 3a, subunit 3 |
| chr14_-_28126346 | 0.03 |
ENSDART00000054062
|
nek12
|
NIMA-related kinase 12 |
| chr10_-_10755962 | 0.03 |
ENSDART00000137797
|
CT027791.1
|
ENSDARG00000093881 |
| chr11_-_25019298 | 0.03 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr9_+_3310926 | 0.03 |
ENSDART00000044128
|
hat1
|
histone acetyltransferase 1 |
| chr2_+_2583038 | 0.03 |
|
|
|
| chr1_+_27115132 | 0.03 |
ENSDART00000166788
|
pspc1
|
paraspeckle component 1 |
| chr2_+_23695713 | 0.03 |
ENSDART00000017034
|
rnf2
|
ring finger protein 2 |
| chr20_-_40357240 | 0.03 |
|
|
|
| chr5_-_9288487 | 0.03 |
ENSDART00000158874
|
ddr2l
|
discoidin domain receptor family, member 2, like |
| chr21_-_1339525 | 0.03 |
|
|
|
| chr1_-_52833426 | 0.03 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr16_-_17678748 | 0.03 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr25_-_35094733 | 0.03 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr14_-_11714513 | 0.03 |
ENSDART00000106654
|
znf711
|
zinc finger protein 711 |
| chr12_+_26931418 | 0.03 |
ENSDART00000152975
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr7_+_36444234 | 0.03 |
|
|
|
| chr20_-_2624028 | 0.03 |
ENSDART00000145335
|
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070841 | inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.2 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.0 | 0.2 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.2 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0051151 | regulation of smooth muscle cell differentiation(GO:0051150) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 0.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |