Project
DANIO-CODE
Navigation
Downloads
Results for vox
Z-value: 0.42
Transcription factors associated with vox
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
vox
|
ENSDARG00000099761 | ventral homeobox |
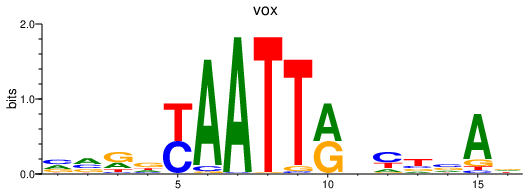
Activity profile of vox motif
Sorted Z-values of vox motif
Network of associatons between targets according to the STRING database.
First level regulatory network of vox
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_22545318 | 0.99 |
ENSDART00000143312
|
zgc:193505
|
zgc:193505 |
| chr1_+_26330186 | 0.60 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| chr4_+_5498355 | 0.60 |
ENSDART00000150785
|
mapk11
|
mitogen-activated protein kinase 11 |
| chr7_+_56405531 | 0.58 |
|
|
|
| chr25_+_19051298 | 0.55 |
ENSDART00000067324
|
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr21_-_22510751 | 0.51 |
ENSDART00000169870
|
myo5b
|
myosin VB |
| chr7_+_6517248 | 0.48 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr21_-_17259886 | 0.47 |
ENSDART00000114877
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr6_-_7563110 | 0.47 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr17_+_23278879 | 0.45 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr7_+_23811321 | 0.42 |
|
|
|
| chr10_+_2952793 | 0.41 |
|
|
|
| chr7_-_45726932 | 0.40 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr16_+_14817799 | 0.39 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr7_+_24770873 | 0.39 |
ENSDART00000165314
|
AL954715.1
|
ENSDARG00000103682 |
| chr5_+_55598181 | 0.38 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr13_-_49695322 | 0.37 |
|
|
|
| chr15_-_15421065 | 0.35 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr23_+_20183765 | 0.34 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr3_-_38642067 | 0.32 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr15_-_1858350 | 0.31 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr7_-_45726974 | 0.31 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr25_+_31457309 | 0.29 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr18_-_44323435 | 0.28 |
ENSDART00000098599
|
si:ch211-151h10.2
|
si:ch211-151h10.2 |
| chr18_+_2203805 | 0.27 |
|
|
|
| chr3_-_15849644 | 0.26 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr5_+_26804344 | 0.25 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr16_-_7525980 | 0.25 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr6_-_39346614 | 0.25 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr24_-_42037243 | 0.24 |
|
|
|
| chr5_-_54026450 | 0.23 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr9_-_30437160 | 0.23 |
ENSDART00000147241
|
BX936337.1
|
ENSDARG00000092870 |
| chr12_-_27032151 | 0.23 |
ENSDART00000153365
|
BX001014.2
|
ENSDARG00000096750 |
| chr15_-_18110169 | 0.23 |
|
|
|
| chr3_+_25992836 | 0.22 |
ENSDART00000010477
|
hsp70.3
|
heat shock cognate 70-kd protein, tandem duplicate 3 |
| chr16_-_24280209 | 0.22 |
ENSDART00000048599
|
rps19
|
ribosomal protein S19 |
| chr16_+_51319421 | 0.22 |
|
|
|
| chr12_+_27032862 | 0.21 |
|
|
|
| chr7_-_25426346 | 0.21 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr2_-_28687065 | 0.21 |
ENSDART00000164657
|
dhcr7
|
7-dehydrocholesterol reductase |
| chr6_-_39278328 | 0.21 |
ENSDART00000148531
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr3_+_47412661 | 0.20 |
ENSDART00000155648
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr9_+_21548069 | 0.19 |
ENSDART00000147619
|
n6amt2
|
N-6 adenine-specific DNA methyltransferase 2 (putative) |
| chr11_-_6442836 | 0.19 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr16_+_54350976 | 0.18 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr23_-_10692678 | 0.18 |
|
|
|
| chr3_-_53467621 | 0.18 |
ENSDART00000032788
|
rdh8a
|
retinol dehydrogenase 8a |
| chr8_+_30912152 | 0.18 |
ENSDART00000171400
|
derl3
|
derlin 3 |
| chr9_-_20562293 | 0.18 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr23_+_13106555 | 0.18 |
|
|
|
| chr1_+_22000499 | 0.17 |
ENSDART00000017413
|
zmynd10
|
zinc finger, MYND-type containing 10 |
| chr10_+_39148251 | 0.17 |
ENSDART00000125986
|
ENSDARG00000055339
|
ENSDARG00000055339 |
| chr10_+_32160464 | 0.17 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr12_-_308929 | 0.17 |
ENSDART00000066579
ENSDART00000171396 |
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr8_-_53508979 | 0.17 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr15_-_5911817 | 0.16 |
|
|
|
| chr15_+_11772499 | 0.16 |
|
|
|
| chr5_+_65754237 | 0.16 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr2_-_58214171 | 0.16 |
ENSDART00000169909
ENSDART00000168590 |
ENSDARG00000103284
|
ENSDARG00000103284 |
| chr15_-_35394535 | 0.16 |
ENSDART00000144153
|
mff
|
mitochondrial fission factor |
| chr5_+_29071526 | 0.16 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr13_+_33578737 | 0.15 |
ENSDART00000161465
|
CABZ01087953.1
|
ENSDARG00000104106 |
| chr6_-_35417797 | 0.15 |
ENSDART00000016586
|
rgs5a
|
regulator of G protein signaling 5a |
| chr12_+_22459371 | 0.15 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr19_+_27133834 | 0.15 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr17_+_42003279 | 0.15 |
ENSDART00000111537
|
kiz
|
kizuna centrosomal protein |
| chr8_-_22252973 | 0.14 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr1_+_26330335 | 0.14 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| KN149726v1_+_1094 | 0.14 |
|
|
|
| chr1_+_49894685 | 0.14 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr4_+_14728248 | 0.13 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr23_+_25378921 | 0.13 |
ENSDART00000143291
|
RPL41
|
ribosomal protein L41 |
| chr14_+_10984191 | 0.13 |
ENSDART00000113567
|
rlim
|
ring finger protein, LIM domain interacting |
| chr22_+_16282153 | 0.12 |
ENSDART00000162685
ENSDART00000105678 |
lrrc39
|
leucine rich repeat containing 39 |
| chr23_-_7740845 | 0.11 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr12_-_309047 | 0.11 |
ENSDART00000066579
ENSDART00000171396 |
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr2_+_22708278 | 0.11 |
ENSDART00000163172
|
hs2st1a
|
heparan sulfate 2-O-sulfotransferase 1a |
| chr16_+_17808801 | 0.11 |
ENSDART00000149596
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr23_+_35614706 | 0.10 |
ENSDART00000123518
ENSDART00000113297 |
tuba1b
|
tubulin, alpha 1b |
| chr8_+_5222065 | 0.10 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr3_+_31047152 | 0.10 |
|
|
|
| chr5_-_64201886 | 0.10 |
ENSDART00000145819
|
krt1-c5
|
keratin, type 1, gene c5 |
| chr21_+_6829352 | 0.10 |
ENSDART00000146371
|
olfm1b
|
olfactomedin 1b |
| chr1_-_39622531 | 0.10 |
|
|
|
| chr3_-_16569378 | 0.09 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr6_+_15635738 | 0.09 |
ENSDART00000128939
|
iqca1
|
IQ motif containing with AAA domain 1 |
| chr5_-_33156615 | 0.09 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr8_-_39788989 | 0.09 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr17_+_44583892 | 0.09 |
ENSDART00000156625
|
pgfb
|
placental growth factor b |
| chr13_-_51409167 | 0.08 |
ENSDART00000174550
|
CABZ01090417.1
|
ENSDARG00000108080 |
| chr25_+_4909062 | 0.08 |
ENSDART00000170049
|
parvb
|
parvin, beta |
| chr11_+_38013238 | 0.07 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr14_-_3836967 | 0.07 |
ENSDART00000055817
|
ENSDARG00000038270
|
ENSDARG00000038270 |
| chr21_+_6829161 | 0.07 |
ENSDART00000037265
|
olfm1b
|
olfactomedin 1b |
| chr25_-_18855316 | 0.07 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr7_+_24762755 | 0.07 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr9_-_12623451 | 0.06 |
ENSDART00000142024
|
tra2b
|
transformer 2 beta homolog |
| chr14_+_26424717 | 0.06 |
|
|
|
| chr5_+_41396720 | 0.06 |
ENSDART00000097580
|
pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr23_+_36016450 | 0.06 |
ENSDART00000103035
|
hoxc6a
|
homeobox C6a |
| chr4_+_14728603 | 0.05 |
ENSDART00000131666
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr19_+_46567459 | 0.05 |
ENSDART00000164055
|
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr19_-_42759951 | 0.05 |
|
|
|
| chr17_+_50622887 | 0.05 |
ENSDART00000134126
|
fermt2
|
fermitin family member 2 |
| KN150030v1_-_22642 | 0.05 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr25_-_13737344 | 0.05 |
|
|
|
| chr7_+_65824459 | 0.05 |
ENSDART00000098259
ENSDART00000051823 ENSDART00000104523 |
arntl1b
|
aryl hydrocarbon receptor nuclear translocator-like 1b |
| chr16_+_17808913 | 0.04 |
ENSDART00000149596
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr3_-_53467536 | 0.04 |
ENSDART00000032788
|
rdh8a
|
retinol dehydrogenase 8a |
| chr12_-_18997267 | 0.03 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr8_+_17042976 | 0.03 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr10_+_37193690 | 0.03 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr6_+_15635696 | 0.02 |
ENSDART00000128939
|
iqca1
|
IQ motif containing with AAA domain 1 |
| chr4_+_75574289 | 0.02 |
ENSDART00000174312
|
CU467646.4
|
ENSDARG00000099945 |
| chr20_-_15029379 | 0.02 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| KN149711v1_-_7827 | 0.02 |
ENSDART00000171410
|
ENSDARG00000104905
|
ENSDARG00000104905 |
| chr15_-_46439102 | 0.02 |
ENSDART00000125211
|
ENSDARG00000087950
|
ENSDARG00000087950 |
| chr24_-_14447136 | 0.01 |
|
|
|
| chr2_-_22708324 | 0.01 |
|
|
|
| chr23_-_34983136 | 0.01 |
|
|
|
| chr13_+_44720106 | 0.01 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr10_+_11306969 | 0.00 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr12_+_22459218 | 0.00 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr19_-_10405099 | 0.00 |
ENSDART00000023571
|
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.2 | GO:0002280 | monocyte activation involved in immune response(GO:0002280) negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0032965 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.0 | 0.3 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.2 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.7 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.3 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.2 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.5 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0097546 | ciliary base(GO:0097546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.2 | GO:0003874 | 6-pyruvoyltetrahydropterin synthase activity(GO:0003874) |
| 0.1 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME BASE EXCISION REPAIR | Genes involved in Base Excision Repair |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |