Project
DANIO-CODE
Navigation
Downloads
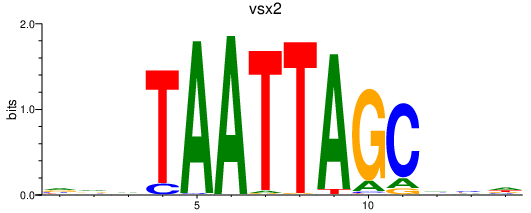
Results for vsx2
Z-value: 0.24
Transcription factors associated with vsx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
vsx2
|
ENSDARG00000005574 | visual system homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vsx2 | dr10_dc_chr17_-_31642673_31642702 | 0.38 | 1.4e-01 | Click! |
Activity profile of vsx2 motif
Sorted Z-values of vsx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of vsx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_46145286 | 0.28 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr1_+_55077656 | 0.26 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr3_-_42649955 | 0.25 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr11_-_37158131 | 0.25 |
ENSDART00000160911
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr5_-_64149931 | 0.22 |
ENSDART00000144816
|
lix1
|
limb and CNS expressed 1 |
| chr7_-_68229145 | 0.19 |
|
|
|
| chr13_+_3533318 | 0.17 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr13_-_49695322 | 0.16 |
|
|
|
| chr3_-_42649907 | 0.16 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr23_+_28656263 | 0.16 |
ENSDART00000020296
|
nadl1.2
|
neural adhesion molecule L1.2 |
| chr23_+_28804842 | 0.15 |
ENSDART00000047378
|
sst3
|
somatostatin 3 |
| chr13_+_3533426 | 0.15 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr3_-_8608620 | 0.14 |
|
|
|
| chr19_+_22478256 | 0.14 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr11_-_6442836 | 0.13 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr1_+_55077417 | 0.12 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr7_-_68229018 | 0.12 |
|
|
|
| chr21_-_13565401 | 0.12 |
ENSDART00000151547
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr24_-_23174888 | 0.11 |
ENSDART00000112256
|
zfhx4
|
zinc finger homeobox 4 |
| chr7_+_36627685 | 0.11 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr13_-_44492897 | 0.11 |
|
|
|
| chr1_+_8605984 | 0.10 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr24_-_23175007 | 0.10 |
ENSDART00000112256
|
zfhx4
|
zinc finger homeobox 4 |
| chr18_+_5899355 | 0.10 |
|
|
|
| chr14_+_23419894 | 0.09 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr16_-_25620142 | 0.09 |
ENSDART00000077420
|
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr3_-_35928594 | 0.09 |
|
|
|
| chr9_-_23340795 | 0.09 |
ENSDART00000133017
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr20_+_26012369 | 0.09 |
ENSDART00000146004
|
ttbk2b
|
tau tubulin kinase 2b |
| chr24_-_14447136 | 0.08 |
|
|
|
| chr5_-_64149894 | 0.08 |
ENSDART00000144816
|
lix1
|
limb and CNS expressed 1 |
| chr24_-_21778717 | 0.07 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr4_+_11465367 | 0.07 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr2_-_27065203 | 0.07 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr12_+_30252424 | 0.07 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr13_+_19191645 | 0.07 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr4_+_3970874 | 0.06 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr9_-_50304120 | 0.06 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr4_+_2215426 | 0.06 |
ENSDART00000168370
|
frs2a
|
fibroblast growth factor receptor substrate 2a |
| chr11_+_18710724 | 0.06 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr10_+_2952793 | 0.05 |
|
|
|
| chr7_-_29861232 | 0.05 |
|
|
|
| chr23_-_24556810 | 0.05 |
ENSDART00000109248
|
spen
|
spen family transcriptional repressor |
| chr13_+_30401664 | 0.04 |
|
|
|
| chr1_-_18709814 | 0.04 |
ENSDART00000145224
|
rbm47
|
RNA binding motif protein 47 |
| chr21_+_28921734 | 0.04 |
ENSDART00000166575
|
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr23_+_17220363 | 0.04 |
ENSDART00000143420
|
BX927275.2
|
ENSDARG00000095017 |
| chr24_-_24859334 | 0.04 |
ENSDART00000080997
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr15_-_20189405 | 0.04 |
ENSDART00000152355
|
med13b
|
mediator complex subunit 13b |
| chr5_+_4906729 | 0.04 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr18_+_7680416 | 0.03 |
ENSDART00000132369
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr24_-_24859135 | 0.03 |
ENSDART00000136860
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr14_+_21522672 | 0.03 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr24_+_40292617 | 0.03 |
|
|
|
| chr11_+_43924640 | 0.02 |
ENSDART00000179206
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr20_+_35176069 | 0.02 |
|
|
|
| chr11_-_3846064 | 0.02 |
ENSDART00000161426
|
gata2a
|
GATA binding protein 2a |
| chr5_+_4906690 | 0.01 |
ENSDART00000124022
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr7_-_29861188 | 0.01 |
|
|
|
| chr11_-_29520809 | 0.01 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr14_+_23419864 | 0.00 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |