Project
DANIO-CODE
Navigation
Downloads
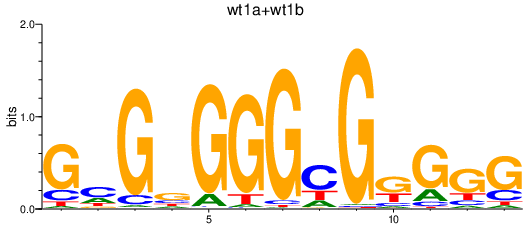
Results for wt1a+wt1b
Z-value: 2.74
Transcription factors associated with wt1a+wt1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
wt1b
|
ENSDARG00000007990 | WT1 transcription factor b |
|
wt1a
|
ENSDARG00000031420 | WT1 transcription factor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| wt1a | dr10_dc_chr25_-_15117843_15117894 | 0.91 | 1.4e-06 | Click! |
| wt1b | dr10_dc_chr18_-_45620179_45620192 | 0.83 | 8.3e-05 | Click! |
Activity profile of wt1a+wt1b motif
Sorted Z-values of wt1a+wt1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of wt1a+wt1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_46768482 | 9.72 |
ENSDART00000015046
|
fstl1a
|
follistatin-like 1a |
| chr14_-_943860 | 7.35 |
ENSDART00000010773
|
acsl1b
|
acyl-CoA synthetase long-chain family member 1b |
| chr19_+_2013850 | 7.15 |
ENSDART00000048850
|
sh3bp5a
|
SH3-domain binding protein 5a (BTK-associated) |
| chr1_-_50831155 | 6.92 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr14_+_21531709 | 6.88 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr9_-_39195468 | 6.20 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr3_+_16115708 | 6.03 |
ENSDART00000122519
|
st8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr1_+_29054033 | 6.00 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr3_+_32394653 | 5.94 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr8_-_53508979 | 5.92 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr23_+_45020761 | 5.86 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr5_+_6054781 | 5.77 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr14_+_41805073 | 5.73 |
ENSDART00000074362
|
pcdh18b
|
protocadherin 18b |
| chr1_-_53247258 | 5.71 |
ENSDART00000007732
|
capn9
|
calpain 9 |
| chr18_-_15404998 | 5.52 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr6_-_39315024 | 5.25 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr16_-_46799406 | 5.17 |
ENSDART00000115244
|
MEX3A
|
mex-3 RNA binding family member A |
| chr12_+_9842842 | 5.08 |
ENSDART00000055019
|
ndufa4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 |
| chr17_+_30687093 | 5.02 |
ENSDART00000179434
|
apoba
|
apolipoprotein Ba |
| chr13_+_25298383 | 4.99 |
|
|
|
| chr4_-_76620869 | 4.95 |
|
|
|
| chr1_-_416138 | 4.94 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr16_-_53896201 | 4.93 |
ENSDART00000179533
|
fzd1
|
frizzled class receptor 1 |
| KN149909v1_+_1319 | 4.85 |
|
|
|
| chr15_+_125263 | 4.84 |
|
|
|
| chr14_-_46359062 | 4.83 |
ENSDART00000090844
|
zgc:153018
|
zgc:153018 |
| chr10_-_28949111 | 4.82 |
ENSDART00000030138
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr17_-_124685 | 4.71 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr6_+_6333766 | 4.63 |
ENSDART00000140827
|
bcl11ab
|
B-cell CLL/lymphoma 11Ab |
| chr10_+_9052152 | 4.51 |
ENSDART00000139466
|
itga2.2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 2 |
| chr13_+_22350043 | 4.43 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr18_-_15405161 | 4.41 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr10_+_4094568 | 4.35 |
ENSDART00000114533
|
mn1a
|
meningioma 1a |
| chr16_-_31959111 | 4.34 |
ENSDART00000135669
|
rbfox1l
|
RNA binding protein, fox-1 homolog (C. elegans) 1-like |
| chr14_+_41804998 | 4.33 |
ENSDART00000074362
|
pcdh18b
|
protocadherin 18b |
| chr18_+_12207888 | 4.33 |
|
|
|
| chr19_+_34114412 | 4.32 |
ENSDART00000162517
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr23_+_2597825 | 4.30 |
ENSDART00000159039
|
ENSDARG00000089986
|
ENSDARG00000089986 |
| chr12_-_954375 | 4.23 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr8_+_54171992 | 4.15 |
ENSDART00000122692
|
CABZ01112317.1
|
ENSDARG00000086057 |
| chr20_-_158899 | 4.13 |
ENSDART00000131635
|
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr2_-_39710140 | 4.12 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr3_+_49277125 | 4.10 |
ENSDART00000161724
|
gas7a
|
growth arrest-specific 7a |
| chr6_-_22504772 | 4.06 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr16_+_1227270 | 4.02 |
|
|
|
| chr4_+_1750689 | 3.98 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr23_-_7889744 | 3.96 |
ENSDART00000164117
|
myt1b
|
myelin transcription factor 1b |
| chr10_+_10830549 | 3.94 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr13_-_508202 | 3.92 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr7_-_50827308 | 3.91 |
ENSDART00000121574
|
col4a6
|
collagen, type IV, alpha 6 |
| chr6_-_2294751 | 3.87 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr2_+_24649130 | 3.85 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr4_-_76594677 | 3.84 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr3_+_14238188 | 3.82 |
ENSDART00000165452
ENSDART00000171726 |
tmem56b
|
transmembrane protein 56b |
| chr24_-_4418454 | 3.78 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr7_-_26165200 | 3.77 |
ENSDART00000123395
|
her8a
|
hairy-related 8a |
| chr9_-_30553051 | 3.74 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr7_-_54407681 | 3.69 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr21_+_28921734 | 3.64 |
ENSDART00000166575
|
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr10_+_32617430 | 3.63 |
ENSDART00000109029
|
map6a
|
microtubule-associated protein 6a |
| chr19_-_32214978 | 3.58 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr7_+_73408688 | 3.57 |
ENSDART00000159745
|
PCP4L1
|
Purkinje cell protein 4 like 1 |
| KN149874v1_+_3991 | 3.55 |
|
|
|
| chr14_+_241493 | 3.53 |
ENSDART00000002232
|
msx1a
|
muscle segment homeobox 1a |
| chr3_+_37645350 | 3.53 |
ENSDART00000134524
|
rps11
|
ribosomal protein S11 |
| chr5_+_71367929 | 3.51 |
ENSDART00000149910
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr16_+_5377008 | 3.46 |
ENSDART00000133972
|
plecb
|
plectin b |
| chr20_-_9593191 | 3.42 |
ENSDART00000014168
|
zfp36l1b
|
zinc finger protein 36, C3H type-like 1b |
| chr3_+_5665300 | 3.40 |
ENSDART00000101807
|
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr11_-_32461160 | 3.39 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr9_-_33916709 | 3.37 |
ENSDART00000028225
|
mao
|
monoamine oxidase |
| chr17_+_11353143 | 3.35 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr14_-_6901415 | 3.33 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr16_-_22218191 | 3.33 |
ENSDART00000140175
|
si:dkey-71b5.3
|
si:dkey-71b5.3 |
| chr13_-_31304614 | 3.32 |
|
|
|
| chr10_+_10252074 | 3.32 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr19_+_31046291 | 3.27 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr17_+_8166167 | 3.27 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr11_+_10557530 | 3.24 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr25_-_340711 | 3.23 |
ENSDART00000178866
|
CU929174.1
|
ENSDARG00000107546 |
| chr25_+_4918339 | 3.21 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr18_+_5456221 | 3.21 |
|
|
|
| chr19_-_19290260 | 3.21 |
|
|
|
| chr19_+_32183937 | 3.21 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr6_+_39373026 | 3.20 |
ENSDART00000157165
|
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr11_+_6809190 | 3.18 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr5_+_42312784 | 3.16 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr23_-_30858769 | 3.16 |
ENSDART00000131285
|
myt1a
|
myelin transcription factor 1a |
| chr17_+_1610578 | 3.16 |
ENSDART00000082101
|
ppp2r5ca
|
protein phosphatase 2, regulatory subunit B', gamma a |
| chr19_-_617298 | 3.15 |
ENSDART00000171387
|
cyp51
|
cytochrome P450, family 51 |
| chr2_-_58111727 | 3.14 |
ENSDART00000004431
ENSDART00000166845 ENSDART00000163999 |
epb41l3b
|
erythrocyte membrane protein band 4.1-like 3b |
| chr13_+_13550656 | 3.14 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| KN149874v1_+_3898 | 3.13 |
|
|
|
| chr20_+_13998066 | 3.12 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr8_+_20125687 | 3.11 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr11_+_11735899 | 3.09 |
ENSDART00000003891
|
jupa
|
junction plakoglobin a |
| chr23_-_624534 | 3.08 |
ENSDART00000132175
|
nadl1.1
|
neural adhesion molecule L1.1 |
| chr4_-_22751641 | 3.08 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr25_+_24152717 | 3.07 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr5_-_71505131 | 3.06 |
ENSDART00000037691
|
tbx5a
|
T-box 5a |
| chr16_-_41994015 | 3.06 |
|
|
|
| chr5_-_55843553 | 3.05 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr12_+_689413 | 3.00 |
ENSDART00000174804
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr7_-_54407461 | 2.91 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr17_-_31678247 | 2.91 |
ENSDART00000143090
|
lin52
|
lin-52 DREAM MuvB core complex component |
| chr2_+_33399405 | 2.90 |
ENSDART00000137207
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_-_44330405 | 2.90 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr21_-_39013447 | 2.88 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr12_-_47699958 | 2.88 |
ENSDART00000171932
ENSDART00000168165 ENSDART00000161985 |
HHEX
|
hematopoietically expressed homeobox |
| chr14_-_2682064 | 2.85 |
ENSDART00000161677
|
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr17_+_24596687 | 2.84 |
ENSDART00000064739
|
rpl13a
|
ribosomal protein L13a |
| KN150131v1_+_59098 | 2.84 |
ENSDART00000175590
|
CABZ01064941.1
|
ENSDARG00000106550 |
| chr12_-_268910 | 2.84 |
ENSDART00000045071
|
foxk2
|
forkhead box K2 |
| KN149987v1_-_29629 | 2.83 |
ENSDART00000159555
ENSDART00000168161 |
ENSDARG00000103311
|
ENSDARG00000103311 |
| chr15_-_47264693 | 2.82 |
|
|
|
| chr16_-_29229687 | 2.78 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr8_-_15071283 | 2.75 |
|
|
|
| chr2_-_3317258 | 2.75 |
ENSDART00000167944
|
wnt3a
|
wingless-type MMTV integration site family, member 3A |
| chr15_-_14616083 | 2.74 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr5_-_13826859 | 2.73 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr10_-_1690605 | 2.72 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr14_-_46359248 | 2.71 |
ENSDART00000090844
|
zgc:153018
|
zgc:153018 |
| chr15_-_1225595 | 2.69 |
|
|
|
| chr23_-_2573949 | 2.66 |
ENSDART00000123322
|
snai1b
|
snail family zinc finger 1b |
| chr10_+_38568464 | 2.63 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr11_-_2107054 | 2.61 |
ENSDART00000173031
|
hoxc6b
|
homeobox C6b |
| chr12_+_41016238 | 2.61 |
ENSDART00000170976
|
kif5bb
|
kinesin family member 5B, b |
| chr8_+_8127635 | 2.59 |
ENSDART00000147940
|
plxnb3
|
plexin B3 |
| chr2_+_43619752 | 2.57 |
ENSDART00000142078
ENSDART00000098265 ENSDART00000098267 |
nrp1b
|
neuropilin 1b |
| chr12_-_35590747 | 2.55 |
|
|
|
| chr23_+_28804842 | 2.52 |
ENSDART00000047378
|
sst3
|
somatostatin 3 |
| chr20_+_23392782 | 2.52 |
|
|
|
| chr1_-_38189932 | 2.51 |
ENSDART00000136102
|
vegfc
|
vascular endothelial growth factor c |
| chr23_-_7283514 | 2.51 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr10_+_1812271 | 2.50 |
|
|
|
| chr8_-_52952226 | 2.47 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr19_+_1814994 | 2.45 |
|
|
|
| chr11_-_232269 | 2.44 |
ENSDART00000172818
|
si:ch1073-456m8.1
|
si:ch1073-456m8.1 |
| chr15_-_19314712 | 2.44 |
ENSDART00000092705
|
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr2_-_44429891 | 2.42 |
ENSDART00000163040
ENSDART00000056372 ENSDART00000109251 ENSDART00000166923 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr2_-_42784609 | 2.40 |
|
|
|
| chr3_+_16115532 | 2.40 |
|
|
|
| chr1_-_50513645 | 2.39 |
ENSDART00000047851
|
jag1a
|
jagged 1a |
| chr16_+_25845432 | 2.39 |
|
|
|
| chr24_+_3296536 | 2.39 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr12_-_48278273 | 2.37 |
|
|
|
| chr5_+_8843643 | 2.37 |
ENSDART00000149417
ENSDART00000061616 |
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr24_-_3273409 | 2.36 |
|
|
|
| chr8_-_2054323 | 2.34 |
ENSDART00000123804
|
cltcl1
|
clathrin, heavy chain-like 1 |
| chr9_-_24602136 | 2.31 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr14_-_9216303 | 2.31 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr10_-_1645 | 2.29 |
|
|
|
| chr23_+_27985224 | 2.29 |
ENSDART00000171859
|
ENSDARG00000100606
|
ENSDARG00000100606 |
| chr2_-_9848848 | 2.28 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr4_-_72603830 | 2.25 |
ENSDART00000168547
|
si:cabz01071912.2
|
si:cabz01071912.2 |
| chr19_+_12066023 | 2.23 |
ENSDART00000130537
|
spag1a
|
sperm associated antigen 1a |
| chr1_-_50513873 | 2.22 |
ENSDART00000137172
|
jag1a
|
jagged 1a |
| chr10_+_10393377 | 2.20 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr5_+_26925238 | 2.16 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr24_+_38783264 | 2.16 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr14_+_23986703 | 2.14 |
ENSDART00000079164
|
klhl3
|
kelch-like family member 3 |
| chr18_-_15498798 | 2.13 |
ENSDART00000109410
|
endouc
|
endonuclease, polyU-specific C |
| chr1_-_13547500 | 2.10 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr1_+_54231447 | 2.10 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr8_+_6533379 | 2.09 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr15_-_47264619 | 2.07 |
|
|
|
| chr14_-_12531063 | 2.07 |
ENSDART00000165004
ENSDART00000043180 |
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr21_+_13764828 | 2.07 |
ENSDART00000171306
|
stxbp1a
|
syntaxin binding protein 1a |
| chr23_+_26995808 | 2.06 |
|
|
|
| chr7_+_31567166 | 2.05 |
ENSDART00000099785
ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr5_+_11340748 | 2.05 |
ENSDART00000159248
|
tesca
|
tescalcin a |
| chr16_+_5778398 | 2.03 |
ENSDART00000011166
ENSDART00000126445 |
zgc:158689
|
zgc:158689 |
| chr20_+_26408687 | 2.03 |
|
|
|
| chr3_-_5318289 | 2.02 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr5_+_22465722 | 2.01 |
ENSDART00000145477
ENSDART00000089992 |
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr10_+_34372205 | 2.00 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr5_+_36014029 | 1.99 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr3_+_32360862 | 1.98 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr20_+_26981663 | 1.97 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr16_-_543957 | 1.96 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr21_+_11046459 | 1.95 |
ENSDART00000169024
|
agxt2
|
alanine--glyoxylate aminotransferase 2 |
| chr18_-_15498733 | 1.95 |
ENSDART00000109410
|
endouc
|
endonuclease, polyU-specific C |
| KN149978v1_-_10669 | 1.95 |
ENSDART00000175723
|
CABZ01068178.1
|
ENSDARG00000105971 |
| chr6_+_55091896 | 1.93 |
|
|
|
| chr12_-_308929 | 1.93 |
ENSDART00000066579
ENSDART00000171396 |
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr22_-_62082 | 1.92 |
ENSDART00000166010
ENSDART00000125700 |
mrpl20
|
mitochondrial ribosomal protein L20 |
| chr17_+_53224434 | 1.92 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr23_+_33457343 | 1.92 |
ENSDART00000114423
|
rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr8_+_54003190 | 1.91 |
|
|
|
| chr3_-_34417082 | 1.90 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr4_-_76594399 | 1.90 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr16_+_21099550 | 1.89 |
|
|
|
| chr10_+_45499009 | 1.89 |
ENSDART00000166085
|
ppiab
|
peptidylprolyl isomerase Ab (cyclophilin A) |
| chr17_-_43475725 | 1.88 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr13_-_508050 | 1.87 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr8_-_46500948 | 1.87 |
ENSDART00000168384
|
sult1st1
|
sulfotransferase family 1, cytosolic sulfotransferase 1 |
| chr25_-_36886647 | 1.86 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr5_+_26925392 | 1.86 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr21_-_14534576 | 1.86 |
ENSDART00000089967
ENSDART00000132142 |
cacna1bb
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, b |
| chr25_+_35269590 | 1.85 |
ENSDART00000034737
|
cpne8
|
copine VIII |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 9.9 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 1.7 | 6.9 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 1.4 | 4.1 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 1.3 | 3.8 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 1.1 | 4.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.1 | 5.4 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic G1 DNA damage checkpoint(GO:0031571) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 1.0 | 3.1 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 1.0 | 3.1 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) |
| 1.0 | 3.0 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 1.0 | 4.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.9 | 6.0 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.8 | 5.9 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.8 | 2.5 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.8 | 3.2 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.8 | 6.0 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.7 | 5.2 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.7 | 2.9 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.7 | 4.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.7 | 2.0 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.6 | 7.1 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.6 | 2.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.6 | 2.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.6 | 4.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.6 | 2.8 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.5 | 1.6 | GO:1904035 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.5 | 1.6 | GO:2000253 | negative regulation of lipid transport(GO:0032369) endocrine process(GO:0050886) positive regulation of feeding behavior(GO:2000253) |
| 0.5 | 2.0 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.4 | 3.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.4 | 1.7 | GO:0051414 | response to cortisol(GO:0051414) cellular response to cortisol stimulus(GO:0071387) |
| 0.4 | 4.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.4 | 2.6 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.4 | 2.1 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.4 | 6.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.4 | 9.7 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.4 | 5.0 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.4 | 3.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.4 | 4.9 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.4 | 6.3 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.4 | 2.6 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.3 | 6.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.3 | 5.9 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.3 | 3.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.3 | 3.1 | GO:0006956 | complement activation(GO:0006956) |
| 0.3 | 2.5 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.3 | 1.8 | GO:0007343 | egg activation(GO:0007343) |
| 0.3 | 3.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.3 | 0.8 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.3 | 1.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.3 | 1.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 | 7.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.3 | 5.4 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.3 | 1.5 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.2 | 2.7 | GO:0021794 | thalamus development(GO:0021794) |
| 0.2 | 0.9 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 3.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.9 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 1.1 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 0.6 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 2.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 2.7 | GO:0003260 | cardioblast migration(GO:0003260) cardioblast migration to the midline involved in heart field formation(GO:0060975) |
| 0.2 | 1.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 1.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.2 | 5.4 | GO:0007586 | digestion(GO:0007586) |
| 0.2 | 3.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.2 | 1.5 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.2 | 1.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 3.9 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.2 | 1.5 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 9.6 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.2 | 0.5 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.2 | 2.6 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 1.8 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.4 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 7.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 2.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 3.5 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 1.6 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 3.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 2.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 2.2 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 4.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 14.7 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 2.0 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.4 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 2.0 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.1 | 2.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 3.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.6 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.1 | 1.0 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 2.4 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 1.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 3.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 3.7 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.1 | 0.8 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.8 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 | 10.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 7.4 | GO:0010721 | negative regulation of cell development(GO:0010721) |
| 0.1 | 0.9 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 1.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.1 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 3.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 4.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 1.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.7 | GO:0072102 | glomerulus morphogenesis(GO:0072102) |
| 0.1 | 4.9 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 1.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 2.9 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.1 | 0.5 | GO:0015961 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 2.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.1 | 1.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 3.7 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 2.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.9 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 1.2 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 1.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.4 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 2.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 1.3 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.6 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 3.3 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.7 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 1.3 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 2.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.3 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 3.4 | GO:0060548 | negative regulation of cell death(GO:0060548) |
| 0.0 | 1.2 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.9 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 6.0 | GO:0007420 | brain development(GO:0007420) |
| 0.0 | 3.0 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 0.4 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.7 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.9 | GO:0043067 | regulation of programmed cell death(GO:0043067) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | GO:0042627 | chylomicron(GO:0042627) |
| 1.0 | 3.9 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.7 | 5.9 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.6 | 2.9 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.5 | 3.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 7.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.4 | 1.7 | GO:0016586 | RSC complex(GO:0016586) |
| 0.4 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 2.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.4 | 4.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 1.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.4 | 2.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.3 | 3.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 3.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 0.8 | GO:1990879 | CST complex(GO:1990879) |
| 0.3 | 2.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 2.3 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 1.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 3.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 4.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 3.7 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.2 | 4.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 7.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 6.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 5.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 2.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 4.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 4.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 3.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) calcium channel complex(GO:0034704) |
| 0.1 | 4.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 2.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 9.5 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 2.8 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 0.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 4.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 42.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 22.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 5.5 | GO:0099503 | secretory vesicle(GO:0099503) |
| 0.0 | 1.5 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 3.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 2.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 3.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.3 | GO:0005840 | ribosome(GO:0005840) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.0 | 3.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.8 | 4.1 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.8 | 4.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.8 | 3.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.7 | 7.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.7 | 5.0 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.7 | 6.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.6 | 1.7 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.6 | 2.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.5 | 6.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 3.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 1.4 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.5 | 1.4 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.4 | 4.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.4 | 2.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.4 | 4.2 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.4 | 3.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 2.9 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.4 | 1.6 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.4 | 3.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 7.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 1.8 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 5.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 5.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 5.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 2.9 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 3.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 3.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 5.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 2.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 0.9 | GO:0003874 | 6-pyruvoyltetrahydropterin synthase activity(GO:0003874) |
| 0.3 | 0.8 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.3 | 1.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 1.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 7.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.2 | 2.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 4.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 0.6 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 1.2 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.2 | 1.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 0.5 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.2 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 4.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 3.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 2.0 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.2 | 0.8 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.5 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 3.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 4.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 2.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 7.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 4.2 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.1 | 7.3 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 0.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 3.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 5.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 2.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 3.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 5.9 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 3.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 2.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 5.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 9.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 1.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.3 | GO:0030165 | calcium-transporting ATPase activity(GO:0005388) PDZ domain binding(GO:0030165) |
| 0.1 | 1.7 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.9 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 4.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 12.3 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 1.7 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 2.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 25.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.4 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 3.0 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 45.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 7.7 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 4.2 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) chemorepellent activity(GO:0045499) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 5.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 3.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 9.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 4.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.7 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 4.3 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 1.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.6 | GO:0015267 | channel activity(GO:0015267) passive transmembrane transporter activity(GO:0022803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.5 | 5.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 3.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 10.8 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 6.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 1.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 5.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 1.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 3.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 3.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.8 | 3.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.7 | 2.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.6 | 2.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 2.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.5 | 5.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.4 | 4.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 2.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 3.0 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.3 | 6.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 2.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 9.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 3.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 2.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 3.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 5.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 4.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 7.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.9 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 2.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 3.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 5.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 5.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.9 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |