Project
DANIO-CODE
Navigation
Downloads
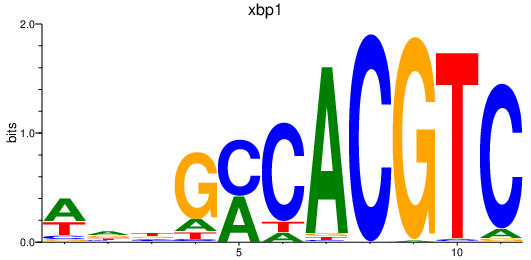
Results for xbp1
Z-value: 1.00
Transcription factors associated with xbp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
xbp1
|
ENSDARG00000035622 | X-box binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| xbp1 | dr10_dc_chr5_-_15448501_15448750 | 0.62 | 9.8e-03 | Click! |
Activity profile of xbp1 motif
Sorted Z-values of xbp1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of xbp1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_10609580 | 2.42 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr13_+_13814142 | 2.09 |
ENSDART00000142997
|
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr11_+_30400284 | 2.09 |
ENSDART00000169833
|
gb:eh507706
|
expressed sequence EH507706 |
| chr10_-_34971985 | 1.86 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr1_-_8801877 | 1.71 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr8_-_32376710 | 1.70 |
ENSDART00000098850
|
lipg
|
lipase, endothelial |
| chr15_+_36075206 | 1.55 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr21_+_10609341 | 1.50 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr1_-_22617455 | 1.48 |
ENSDART00000137567
|
smim14
|
small integral membrane protein 14 |
| chr19_-_5186692 | 1.45 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr7_+_17848688 | 1.36 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr17_+_15205550 | 1.35 |
ENSDART00000058351
|
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr15_+_36075070 | 1.31 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr18_+_6747156 | 1.27 |
ENSDART00000111343
|
lmf2a
|
lipase maturation factor 2a |
| chr14_-_35513309 | 1.26 |
ENSDART00000173343
|
pdgfc
|
platelet derived growth factor c |
| chr5_-_28368834 | 1.26 |
|
|
|
| chr22_+_1500412 | 1.26 |
ENSDART00000160406
|
si:ch211-255f4.5
|
si:ch211-255f4.5 |
| chr5_+_24039974 | 1.25 |
ENSDART00000029699
|
atp6v0a2b
|
ATPase, H+ transporting, lysosomal V0 subunit a2b |
| chr18_-_15300493 | 1.21 |
ENSDART00000048206
|
tmem263
|
transmembrane protein 263 |
| chr16_-_34471672 | 1.19 |
ENSDART00000172162
|
CR626886.2
|
ENSDARG00000105308 |
| chr3_-_25246374 | 1.18 |
ENSDART00000077538
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr12_+_30673985 | 1.17 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr6_-_42390995 | 1.11 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr13_-_46627166 | 1.09 |
|
|
|
| chr7_+_45748086 | 1.09 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr18_-_7522063 | 1.08 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr6_+_13676955 | 1.08 |
ENSDART00000043522
|
tmem198b
|
transmembrane protein 198b |
| chr3_+_28729443 | 1.07 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr8_-_14516806 | 1.04 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr13_-_46627216 | 1.02 |
|
|
|
| chr19_+_27056002 | 1.00 |
ENSDART00000175951
|
ints3
|
integrator complex subunit 3 |
| chr9_+_24014550 | 0.99 |
ENSDART00000138470
|
ints6
|
integrator complex subunit 6 |
| chr15_-_29393378 | 0.96 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr3_-_25246452 | 0.95 |
ENSDART00000177576
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr14_-_15777250 | 0.94 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr4_-_765957 | 0.94 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr12_-_35481361 | 0.93 |
ENSDART00000158658
ENSDART00000168958 ENSDART00000162175 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr13_+_35799851 | 0.93 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr19_+_7630777 | 0.91 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr13_-_42410874 | 0.90 |
ENSDART00000146843
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr25_-_2908684 | 0.87 |
ENSDART00000073618
|
scamp2
|
secretory carrier membrane protein 2 |
| chr12_-_2834771 | 0.86 |
ENSDART00000114854
ENSDART00000163759 |
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr15_-_29393320 | 0.84 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr16_-_34471544 | 0.84 |
ENSDART00000172162
|
CR626886.2
|
ENSDARG00000105308 |
| chr19_+_27055869 | 0.83 |
ENSDART00000067793
ENSDART00000124755 |
ints3
|
integrator complex subunit 3 |
| chr2_+_21516908 | 0.82 |
ENSDART00000079518
|
dnajc1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr10_+_37229202 | 0.80 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr6_-_32099929 | 0.80 |
ENSDART00000150934
|
alg6
|
asparagine-linked glycosylation 6 (alpha-1,3,-glucosyltransferase) |
| chr1_+_45907424 | 0.77 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr16_-_34471475 | 0.77 |
ENSDART00000172162
|
CR626886.2
|
ENSDARG00000105308 |
| chr12_-_16403282 | 0.76 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr8_+_23840165 | 0.74 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr17_+_24427354 | 0.74 |
ENSDART00000163221
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr1_+_51998406 | 0.73 |
ENSDART00000134658
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr17_-_23689380 | 0.73 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr25_-_36691657 | 0.71 |
ENSDART00000153789
|
rfwd3
|
ring finger and WD repeat domain 3 |
| chr6_+_49724436 | 0.70 |
ENSDART00000154738
|
stx16
|
syntaxin 16 |
| chr1_-_52642562 | 0.69 |
ENSDART00000150579
|
znf330
|
zinc finger protein 330 |
| chr7_+_32422659 | 0.69 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr22_-_11049321 | 0.68 |
ENSDART00000006927
ENSDART00000170509 |
use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr25_+_3634121 | 0.66 |
ENSDART00000153905
|
thoc5
|
THO complex 5 |
| chr23_-_24299965 | 0.66 |
ENSDART00000109134
|
plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr3_-_33362384 | 0.65 |
ENSDART00000020342
|
sgsm3
|
small G protein signaling modulator 3 |
| chr5_-_8406033 | 0.64 |
ENSDART00000097221
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr1_-_52642446 | 0.63 |
ENSDART00000131520
|
znf330
|
zinc finger protein 330 |
| chr5_-_56293700 | 0.62 |
|
|
|
| chr17_+_24427881 | 0.62 |
ENSDART00000140467
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr19_+_7630609 | 0.61 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr12_+_31668126 | 0.61 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr13_-_44671682 | 0.61 |
ENSDART00000099984
|
glo1
|
glyoxalase 1 |
| chr7_+_31067562 | 0.60 |
ENSDART00000173477
|
casc4
|
cancer susceptibility candidate 4 |
| chr3_+_27639256 | 0.60 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr1_-_8801818 | 0.59 |
ENSDART00000008682
ENSDART00000157814 |
micall2b
|
mical-like 2b |
| chr25_+_26458700 | 0.59 |
|
|
|
| chr2_-_9729035 | 0.58 |
ENSDART00000004398
|
cope
|
coatomer protein complex, subunit epsilon |
| chr5_+_1737449 | 0.57 |
ENSDART00000064088
|
vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr7_+_31067219 | 0.57 |
|
|
|
| chr7_+_31067469 | 0.57 |
ENSDART00000173477
|
casc4
|
cancer susceptibility candidate 4 |
| chr2_-_42643265 | 0.56 |
ENSDART00000009093
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr8_-_22639164 | 0.56 |
|
|
|
| chr13_-_36440277 | 0.56 |
ENSDART00000030133
|
synj2bp
|
synaptojanin 2 binding protein |
| chr3_+_34051311 | 0.55 |
ENSDART00000055252
|
TIMM29
|
translocase of inner mitochondrial membrane 29 |
| chr12_+_31668168 | 0.55 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr6_-_32100075 | 0.54 |
ENSDART00000130627
|
alg6
|
asparagine-linked glycosylation 6 (alpha-1,3,-glucosyltransferase) |
| chr5_-_29916352 | 0.53 |
ENSDART00000014666
|
arcn1a
|
archain 1a |
| chr1_-_52642622 | 0.53 |
ENSDART00000150579
|
znf330
|
zinc finger protein 330 |
| chr20_-_38851375 | 0.52 |
|
|
|
| chr9_+_54736764 | 0.52 |
ENSDART00000148928
|
rbm27
|
RNA binding motif protein 27 |
| chr17_-_24848180 | 0.52 |
ENSDART00000027957
|
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr13_-_42410700 | 0.52 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr10_+_36751832 | 0.52 |
ENSDART00000169015
|
rab6a
|
RAB6A, member RAS oncogene family |
| chr9_+_19812730 | 0.51 |
ENSDART00000110009
|
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr12_-_2834814 | 0.50 |
ENSDART00000114854
ENSDART00000163759 |
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr14_+_8288174 | 0.50 |
ENSDART00000037749
|
stx5a
|
syntaxin 5A |
| chr22_-_800449 | 0.49 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr10_-_44441197 | 0.49 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr8_-_31691921 | 0.48 |
ENSDART00000141799
|
fbxo4
|
F-box protein 4 |
| chr5_+_1737408 | 0.48 |
ENSDART00000064088
|
vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr14_-_26113593 | 0.48 |
ENSDART00000020582
|
tmed9
|
transmembrane p24 trafficking protein 9 |
| chr23_+_24672673 | 0.47 |
ENSDART00000131689
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr8_-_14516930 | 0.46 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr1_-_52642475 | 0.46 |
ENSDART00000131520
|
znf330
|
zinc finger protein 330 |
| chr2_-_9729223 | 0.45 |
ENSDART00000004398
|
cope
|
coatomer protein complex, subunit epsilon |
| chr1_+_28994314 | 0.43 |
ENSDART00000019743
|
tm9sf2
|
transmembrane 9 superfamily member 2 |
| chr9_-_50106512 | 0.43 |
|
|
|
| chr18_+_7595039 | 0.40 |
ENSDART00000062143
|
zgc:77650
|
zgc:77650 |
| chr12_-_35481239 | 0.40 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr6_+_13273286 | 0.39 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr20_-_1170970 | 0.39 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr7_+_31067321 | 0.39 |
|
|
|
| chr5_-_56294194 | 0.37 |
ENSDART00000074400
|
tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr6_+_6621888 | 0.37 |
ENSDART00000143359
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr1_+_51998508 | 0.37 |
ENSDART00000134658
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr18_+_18598885 | 0.37 |
|
|
|
| chr19_+_7630847 | 0.37 |
ENSDART00000151758
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr20_+_22900165 | 0.36 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr25_+_3634007 | 0.36 |
ENSDART00000055845
|
thoc5
|
THO complex 5 |
| chr10_-_44441390 | 0.36 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr9_+_19812539 | 0.34 |
ENSDART00000147662
|
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr4_+_76572863 | 0.34 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr15_-_29393252 | 0.33 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr3_+_34690574 | 0.33 |
|
|
|
| chr17_+_23291331 | 0.33 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr14_-_46477165 | 0.33 |
ENSDART00000173209
ENSDART00000164837 |
ehd1a
|
EH-domain containing 1a |
| chr16_+_12376861 | 0.32 |
ENSDART00000060037
|
ENSDARG00000040971
|
ENSDARG00000040971 |
| chr7_+_45748322 | 0.31 |
ENSDART00000169822
|
ccne1
|
cyclin E1 |
| chr23_+_44069409 | 0.31 |
ENSDART00000149088
|
nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr23_-_21608362 | 0.31 |
ENSDART00000010647
|
rcc2
|
regulator of chromosome condensation 2 |
| chr6_+_17928720 | 0.31 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr1_-_25369230 | 0.30 |
ENSDART00000171752
|
pdcd4a
|
programmed cell death 4a |
| chr24_-_37178907 | 0.29 |
|
|
|
| chr22_+_34726889 | 0.28 |
ENSDART00000082066
|
atpv0e2
|
ATPase, H+ transporting V0 subunit e2 |
| chr11_+_7267389 | 0.28 |
ENSDART00000165536
|
si:ch73-238c9.1
|
si:ch73-238c9.1 |
| chr9_+_35141690 | 0.28 |
ENSDART00000140851
ENSDART00000077800 |
tfdp1a
|
transcription factor Dp-1, a |
| chr10_+_11306969 | 0.28 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr9_-_50106442 | 0.27 |
|
|
|
| chr14_-_15776937 | 0.26 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr3_-_37447038 | 0.26 |
ENSDART00000149258
|
arf2a
|
ADP-ribosylation factor 2a |
| chr22_+_34726990 | 0.25 |
ENSDART00000082066
|
atpv0e2
|
ATPase, H+ transporting V0 subunit e2 |
| chr3_-_34050856 | 0.25 |
ENSDART00000151819
|
yipf2
|
Yip1 domain family, member 2 |
| chr2_-_58411390 | 0.24 |
ENSDART00000177160
|
AL954696.2
|
ENSDARG00000106834 |
| chr16_+_38209634 | 0.24 |
ENSDART00000160332
|
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr20_+_22900302 | 0.24 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr19_-_11320648 | 0.23 |
ENSDART00000148697
|
ssr2
|
signal sequence receptor, beta |
| chr24_+_14070379 | 0.23 |
ENSDART00000004664
|
tram1
|
translocation associated membrane protein 1 |
| chr5_-_39610208 | 0.22 |
ENSDART00000131323
|
si:dkey-193c22.1
|
si:dkey-193c22.1 |
| chr9_+_30301667 | 0.22 |
ENSDART00000013591
|
tfg
|
trk-fused gene |
| chr25_-_3633631 | 0.21 |
ENSDART00000159335
ENSDART00000088077 |
zgc:158398
|
zgc:158398 |
| chr7_+_17848586 | 0.21 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr12_-_16403608 | 0.20 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr10_-_26263954 | 0.20 |
ENSDART00000079194
|
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr19_+_7630730 | 0.19 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr25_+_31746760 | 0.18 |
ENSDART00000162636
|
tjp1b
|
tight junction protein 1b |
| chr2_+_17130843 | 0.18 |
|
|
|
| chr14_-_46476881 | 0.18 |
ENSDART00000173209
ENSDART00000164837 |
ehd1a
|
EH-domain containing 1a |
| chr25_+_34357376 | 0.17 |
ENSDART00000149782
|
CHST6
|
carbohydrate sulfotransferase 6 |
| chr16_+_27289620 | 0.17 |
ENSDART00000137408
|
sec61b
|
Sec61 translocon beta subunit |
| chr15_-_18179431 | 0.17 |
ENSDART00000047902
|
arcn1b
|
archain 1b |
| chr1_-_6989807 | 0.16 |
ENSDART00000013264
|
arglu1b
|
arginine and glutamate rich 1b |
| chr13_-_33527029 | 0.15 |
|
|
|
| chr14_-_233888 | 0.15 |
ENSDART00000051893
|
stx18
|
syntaxin 18 |
| chr6_+_49724271 | 0.15 |
ENSDART00000002693
|
stx16
|
syntaxin 16 |
| chr19_+_19330545 | 0.13 |
ENSDART00000157523
|
slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr18_-_16934756 | 0.12 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr21_+_28687401 | 0.11 |
ENSDART00000138017
|
puraa
|
purine-rich element binding protein Aa |
| chr11_+_7267123 | 0.11 |
ENSDART00000172407
|
si:ch73-238c9.1
|
si:ch73-238c9.1 |
| chr10_-_26263737 | 0.10 |
ENSDART00000079194
|
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr1_-_54453712 | 0.10 |
ENSDART00000074083
|
rab1aa
|
RAB1A, member RAS oncogene family a |
| chr2_+_37194305 | 0.10 |
ENSDART00000176827
ENSDART00000176583 |
copa
|
coatomer protein complex, subunit alpha |
| chr11_+_6421113 | 0.10 |
ENSDART00000016475
|
VMAC
|
vimentin type intermediate filament associated coiled-coil protein |
| chr16_+_53717064 | 0.09 |
ENSDART00000083513
|
sacm1lb
|
SAC1 like phosphatidylinositide phosphatase b |
| chr3_+_34690823 | 0.08 |
|
|
|
| chr15_-_16134595 | 0.07 |
ENSDART00000122099
|
dynll2a
|
dynein, light chain, LC8-type 2a |
| chr2_+_37844454 | 0.06 |
ENSDART00000121768
|
RNaseP_nuc
|
Nuclear RNase P |
| chr9_+_26029780 | 0.06 |
ENSDART00000127135
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr10_-_3427119 | 0.06 |
ENSDART00000037183
|
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr3_+_34690515 | 0.06 |
|
|
|
| chr20_-_449951 | 0.04 |
ENSDART00000009196
|
ufl1
|
UFM1-specific ligase 1 |
| chr4_-_766308 | 0.04 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr3_-_40407003 | 0.04 |
|
|
|
| chr17_-_45036982 | 0.04 |
ENSDART00000155043
|
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr22_+_31110716 | 0.03 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr20_-_23184071 | 0.03 |
ENSDART00000176282
|
CU693368.2
|
ENSDARG00000108718 |
| chr8_+_47228598 | 0.02 |
ENSDART00000075068
|
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr15_+_1569864 | 0.02 |
ENSDART00000056765
|
smc4
|
structural maintenance of chromosomes 4 |
| chr17_+_19610407 | 0.02 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr10_-_8476067 | 0.02 |
ENSDART00000108643
|
tctn1
|
tectonic family member 1 |
| chr13_-_49934345 | 0.02 |
ENSDART00000034541
|
gpatch11
|
G patch domain containing 11 |
| chr4_-_5904787 | 0.02 |
ENSDART00000169439
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr11_+_44909949 | 0.01 |
ENSDART00000173116
ENSDART00000167011 ENSDART00000167226 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr13_+_36469737 | 0.01 |
ENSDART00000164545
|
cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr3_+_3923243 | 0.01 |
ENSDART00000104977
|
pick1
|
protein interacting with prkca 1 |
| chr20_-_1171011 | 0.01 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr19_+_7255491 | 0.01 |
ENSDART00000151220
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr10_+_16207230 | 0.00 |
ENSDART00000065045
|
prrc1
|
proline-rich coiled-coil 1 |
| chr14_-_31226033 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 2.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 0.9 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.3 | 1.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 1.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.3 | 0.8 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.3 | 1.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) vitamin K metabolic process(GO:0042373) |
| 0.2 | 1.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.7 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.2 | 0.9 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.2 | 2.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 3.2 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 1.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.5 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 | 0.7 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 1.8 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 1.3 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 1.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 2.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.8 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 1.2 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 3.3 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.8 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.7 | GO:1903729 | membrane tubulation(GO:0097320) regulation of plasma membrane organization(GO:1903729) |
| 0.0 | 1.0 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.0 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.2 | GO:0090114 | COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.7 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 2.1 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.0 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.3 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.0 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) regulation of endoplasmic reticulum unfolded protein response(GO:1900101) negative regulation of endoplasmic reticulum unfolded protein response(GO:1900102) regulation of IRE1-mediated unfolded protein response(GO:1903894) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.3 | 1.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.4 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 1.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.2 | 1.0 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 1.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.0 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 5.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 1.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 1.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 1.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.6 | GO:0030141 | secretory granule(GO:0030141) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.5 | 1.4 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 2.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.4 | 1.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.3 | 0.9 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.3 | 1.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 1.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.3 | 1.5 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.2 | 0.9 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.2 | 1.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.1 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.4 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 1.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.7 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.6 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 1.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.3 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 0.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 3.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.0 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 2.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.1 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.2 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 1.3 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 1.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 2.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 3.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 4.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 0.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.1 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 2.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |