Project
DANIO-CODE
Navigation
Downloads
Results for ybx1
Z-value: 1.01
Transcription factors associated with ybx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ybx1
|
ENSDARG00000004757 | Y box binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ybx1 | dr10_dc_chr8_-_47161379_47161678 | 0.76 | 5.8e-04 | Click! |
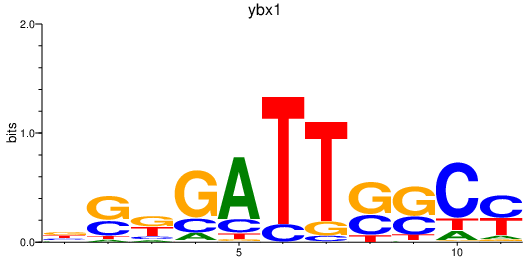
Activity profile of ybx1 motif
Sorted Z-values of ybx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ybx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_2615160 | 4.83 |
ENSDART00000140760
|
e2f7
|
E2F transcription factor 7 |
| chr10_+_39141022 | 3.61 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr4_+_29209923 | 3.51 |
ENSDART00000168709
|
ENSDARG00000104041
|
ENSDARG00000104041 |
| chr10_+_39140943 | 2.25 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr1_+_23385409 | 2.22 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr19_+_12315359 | 2.14 |
ENSDART00000149221
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr4_-_16556116 | 2.11 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr1_+_50277442 | 1.90 |
ENSDART00000150420
|
otx1a
|
orthodenticle homeobox 1a |
| chr16_+_4825019 | 1.85 |
ENSDART00000167665
|
LIN28A
|
lin-28 homolog A |
| chr21_+_5781882 | 1.70 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr18_+_62936 | 1.67 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| KN150670v1_-_72156 | 1.66 |
|
|
|
| chr14_-_40454194 | 1.66 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr14_-_11765407 | 1.64 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr16_+_1338291 | 1.62 |
ENSDART00000149299
|
cers2b
|
ceramide synthase 2b |
| chr11_-_30105047 | 1.60 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr6_+_56163589 | 1.55 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr6_-_50705420 | 1.52 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr21_-_4842528 | 1.45 |
ENSDART00000097796
|
rnf165a
|
ring finger protein 165a |
| chr21_+_19034242 | 1.42 |
ENSDART00000123309
|
nkx6.1
|
NK6 homeobox 1 |
| chr24_-_39884167 | 1.40 |
ENSDART00000087441
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr5_+_69383382 | 1.38 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr11_+_34788205 | 1.34 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr18_+_22617072 | 1.29 |
ENSDART00000128965
|
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr3_-_6078015 | 1.28 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr13_-_2259405 | 1.27 |
ENSDART00000166456
|
elovl5
|
ELOVL fatty acid elongase 5 |
| chr10_+_9325719 | 1.26 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr19_+_32579358 | 1.26 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr16_+_46728964 | 1.24 |
ENSDART00000163571
|
rab25b
|
RAB25, member RAS oncogene family b |
| chr10_+_10830549 | 1.23 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr19_+_12315183 | 1.23 |
ENSDART00000149221
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr3_+_21925465 | 1.21 |
|
|
|
| chr11_-_15162109 | 1.15 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr23_+_44951868 | 1.15 |
|
|
|
| chr13_-_30515486 | 1.15 |
ENSDART00000139607
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr15_+_1187249 | 1.13 |
ENSDART00000152638
ENSDART00000152466 |
mlf1
|
myeloid leukemia factor 1 |
| chr6_+_251026 | 1.09 |
ENSDART00000042970
|
atf4a
|
activating transcription factor 4a |
| chr4_+_30012767 | 1.09 |
ENSDART00000150383
|
znf1128
|
zinc finger protein 1128 |
| chr18_+_5456221 | 1.07 |
|
|
|
| chr19_-_1905372 | 1.04 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr25_+_34515221 | 1.04 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr6_+_3549861 | 1.01 |
ENSDART00000170781
|
phospho2
|
phosphatase, orphan 2 |
| chr23_+_44817648 | 0.99 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr23_+_6861489 | 0.99 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr23_-_41756325 | 0.98 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr6_+_48138737 | 0.97 |
|
|
|
| chr25_-_3521512 | 0.97 |
ENSDART00000171863
ENSDART00000166363 |
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr7_+_5784612 | 0.95 |
ENSDART00000114327
|
zgc:112234
|
zgc:112234 |
| chr9_+_13149212 | 0.93 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr20_-_53133918 | 0.91 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr20_-_53560663 | 0.89 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr25_+_35855292 | 0.89 |
ENSDART00000152801
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr21_-_26369545 | 0.83 |
ENSDART00000077200
|
eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr16_+_52779257 | 0.82 |
|
|
|
| chr17_-_21179064 | 0.82 |
|
|
|
| chr19_-_15951003 | 0.82 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr3_+_2013046 | 0.80 |
ENSDART00000112979
|
sox10
|
SRY (sex determining region Y)-box 10 |
| chr22_+_10172134 | 0.80 |
ENSDART00000132641
|
pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr20_+_16843502 | 0.78 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr18_-_49121584 | 0.78 |
ENSDART00000174157
|
ENSDARG00000099137
|
ENSDARG00000099137 |
| chr25_+_34546629 | 0.74 |
ENSDART00000133379
|
hist2h3c
|
histone cluster 2, H3c |
| chr16_+_11260802 | 0.74 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr19_+_48499602 | 0.74 |
|
|
|
| chr8_-_4451417 | 0.73 |
ENSDART00000141915
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr19_-_34508557 | 0.73 |
ENSDART00000160495
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr1_-_11707288 | 0.71 |
ENSDART00000134708
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr19_-_10962536 | 0.70 |
ENSDART00000160438
|
psmd4a
|
proteasome 26S subunit, non-ATPase 4a |
| chr20_+_54464026 | 0.68 |
ENSDART00000158810
ENSDART00000161631 ENSDART00000172631 ENSDART00000168924 |
fkbp3
|
FK506 binding protein 3 |
| chr1_+_25378105 | 0.68 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr11_-_40255633 | 0.67 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr6_+_2849460 | 0.67 |
|
|
|
| chr20_+_54463948 | 0.66 |
ENSDART00000158810
ENSDART00000161631 ENSDART00000172631 ENSDART00000168924 |
fkbp3
|
FK506 binding protein 3 |
| chr14_-_9274939 | 0.64 |
|
|
|
| chr15_+_41878670 | 0.63 |
ENSDART00000137434
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr9_+_24891584 | 0.61 |
|
|
|
| chr21_-_797123 | 0.61 |
ENSDART00000144766
|
nars
|
asparaginyl-tRNA synthetase |
| chr22_+_38220826 | 0.60 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr15_-_20795531 | 0.60 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr4_+_13451169 | 0.59 |
ENSDART00000172552
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr20_-_47576935 | 0.59 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr25_-_8475902 | 0.58 |
ENSDART00000176751
|
CU929451.3
|
ENSDARG00000107886 |
| chr3_-_58488929 | 0.58 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
| chr7_-_56533829 | 0.57 |
ENSDART00000010322
|
senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr6_-_48095609 | 0.57 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr1_-_18110990 | 0.55 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr13_+_4312466 | 0.55 |
ENSDART00000156323
|
tb
|
T, brachyury homolog b |
| chr6_-_34974860 | 0.55 |
ENSDART00000113097
|
hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr25_-_13737344 | 0.55 |
|
|
|
| chr17_-_52435859 | 0.55 |
|
|
|
| chr9_-_41072329 | 0.55 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr11_-_18442094 | 0.54 |
ENSDART00000159380
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr25_-_3521317 | 0.53 |
ENSDART00000171863
ENSDART00000166363 |
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr4_+_45204456 | 0.53 |
ENSDART00000158744
|
BX927336.3
|
ENSDARG00000102848 |
| chr6_+_53349584 | 0.50 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr6_+_10636497 | 0.50 |
ENSDART00000041858
|
mtx2
|
metaxin 2 |
| chr5_+_4906729 | 0.50 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr8_-_1154004 | 0.49 |
ENSDART00000149969
ENSDART00000016800 |
znf367
|
zinc finger protein 367 |
| chr4_-_16555765 | 0.49 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr23_+_14834670 | 0.49 |
ENSDART00000143675
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr2_-_39709905 | 0.48 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr11_+_26152136 | 0.48 |
|
|
|
| chr12_-_36191048 | 0.47 |
ENSDART00000177986
|
cep131
|
centrosomal protein 131 |
| chr3_+_62081343 | 0.47 |
ENSDART00000058719
|
znf1124
|
zinc finger protein 1124 |
| chr8_+_47909655 | 0.47 |
|
|
|
| chr19_-_791149 | 0.46 |
ENSDART00000151782
ENSDART00000037515 |
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr1_-_54294303 | 0.45 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr10_+_36082726 | 0.45 |
ENSDART00000165386
|
hmgb1a
|
high mobility group box 1a |
| chr25_+_35762694 | 0.44 |
ENSDART00000136456
ENSDART00000175090 |
CR354435.8
zgc:173552
|
Histone H3.2 zgc:173552 |
| chr23_-_5429419 | 0.44 |
ENSDART00000159105
|
CR936540.3
|
ENSDARG00000101435 |
| chr14_+_16508093 | 0.44 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr7_+_73601671 | 0.42 |
ENSDART00000163075
|
zgc:173552
|
zgc:173552 |
| chr25_+_34557335 | 0.42 |
ENSDART00000089844
|
zgc:113983
|
zgc:113983 |
| chr5_+_2839985 | 0.41 |
|
|
|
| chr14_-_16171165 | 0.41 |
ENSDART00000089021
|
canx
|
calnexin |
| chr16_+_38335706 | 0.40 |
|
|
|
| chr19_+_24340673 | 0.40 |
|
|
|
| chr24_-_41356844 | 0.39 |
ENSDART00000121592
ENSDART00000019975 ENSDART00000166034 |
acvr2b
|
activin A receptor, type IIB |
| chr4_+_36180573 | 0.39 |
ENSDART00000161159
|
ENSDARG00000102102
|
ENSDARG00000102102 |
| chr7_+_67510360 | 0.38 |
ENSDART00000172015
|
cyb5b
|
cytochrome b5 type B |
| chr22_+_37688585 | 0.38 |
ENSDART00000174664
ENSDART00000178200 |
FO704589.1
|
ENSDARG00000108910 |
| chr7_-_24119645 | 0.38 |
ENSDART00000141769
|
ptgr1
|
prostaglandin reductase 1 |
| chr16_-_52761159 | 0.38 |
ENSDART00000111869
|
ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr24_+_36478896 | 0.37 |
|
|
|
| chr1_-_29919280 | 0.37 |
ENSDART00000152175
|
MZT1
|
mitotic spindle organizing protein 1 |
| chr4_-_74789596 | 0.37 |
ENSDART00000174356
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr16_+_5411816 | 0.37 |
|
|
|
| chr19_-_30975279 | 0.37 |
ENSDART00000171006
|
hpcal4
|
hippocalcin like 4 |
| chr5_+_35839198 | 0.37 |
ENSDART00000102973
ENSDART00000103020 |
eda
|
ectodysplasin A |
| chr8_+_8889274 | 0.36 |
ENSDART00000142075
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr16_+_49423377 | 0.36 |
ENSDART00000015694
|
rab5ab
|
RAB5A, member RAS oncogene family, b |
| chr21_-_26453406 | 0.36 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr4_-_9195261 | 0.36 |
ENSDART00000080943
|
hcfc2
|
host cell factor C2 |
| chr7_+_5783441 | 0.35 |
ENSDART00000167099
|
CU459186.5
|
Histone H3.2 |
| chr10_+_6009364 | 0.35 |
ENSDART00000163680
|
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr6_-_25962268 | 0.35 |
ENSDART00000076997
|
lmo4b
|
LIM domain only 4b |
| chr9_+_41072345 | 0.34 |
ENSDART00000141548
|
hibch
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr7_-_6330257 | 0.33 |
ENSDART00000173138
|
zgc:112234
|
zgc:112234 |
| chr8_-_34077790 | 0.33 |
|
|
|
| chr7_-_6308104 | 0.32 |
ENSDART00000173010
|
zgc:112234
|
zgc:112234 |
| chr9_-_2416300 | 0.31 |
|
|
|
| chr2_+_29265967 | 0.31 |
ENSDART00000099157
|
cdh18a
|
cadherin 18, type 2a |
| chr25_-_35037680 | 0.30 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr17_-_31204651 | 0.29 |
ENSDART00000137834
|
pcmtl
|
l-isoaspartyl protein carboxyl methyltransferase, like |
| chr19_+_43434941 | 0.29 |
ENSDART00000038230
|
snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr11_+_43334742 | 0.28 |
|
|
|
| chr10_+_1610794 | 0.28 |
ENSDART00000060946
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr13_-_30515304 | 0.27 |
ENSDART00000139607
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr22_-_16154562 | 0.26 |
ENSDART00000090390
|
vcam1b
|
vascular cell adhesion molecule 1b |
| chr3_-_60985683 | 0.26 |
ENSDART00000133091
|
pvalb5
|
parvalbumin 5 |
| chr2_-_56531858 | 0.26 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr12_+_46944395 | 0.26 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr3_+_15245529 | 0.26 |
|
|
|
| chr22_-_3954917 | 0.25 |
ENSDART00000162523
|
kdm4b
|
lysine (K)-specific demethylase 4B |
| chr6_+_52350428 | 0.25 |
ENSDART00000151612
|
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| KN149926v1_+_1959 | 0.25 |
|
|
|
| chr11_-_42263721 | 0.25 |
ENSDART00000130573
|
atp6ap1la
|
ATPase, H+ transporting, lysosomal accessory protein 1-like a |
| chr18_-_26912259 | 0.23 |
ENSDART00000147735
|
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr25_-_34557204 | 0.23 |
|
|
|
| chr23_-_2027166 | 0.22 |
ENSDART00000127443
|
prdm5
|
PR domain containing 5 |
| chr19_-_43434820 | 0.21 |
ENSDART00000122135
|
zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr8_+_16702533 | 0.21 |
ENSDART00000100698
|
ercc8
|
excision repair cross-complementation group 8 |
| chr22_+_36934624 | 0.21 |
ENSDART00000158293
|
mccc1
|
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr4_-_5238656 | 0.20 |
ENSDART00000050221
|
atp6v1e1b
|
ATPase, H+ transporting, lysosomal, V1 subunit E1b |
| chr18_+_5637588 | 0.20 |
|
|
|
| chr21_+_5597476 | 0.20 |
ENSDART00000161235
ENSDART00000170456 |
shroom3
|
shroom family member 3 |
| chr5_+_23582396 | 0.20 |
ENSDART00000051549
ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr7_+_26278650 | 0.18 |
ENSDART00000141353
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr17_-_43296680 | 0.18 |
ENSDART00000176637
|
EML5
|
echinoderm microtubule associated protein like 5 |
| chr13_+_42476309 | 0.18 |
ENSDART00000133388
|
mlh1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) |
| chr15_-_19314712 | 0.18 |
ENSDART00000092705
|
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr16_+_68014 | 0.17 |
ENSDART00000159652
|
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr4_-_18660286 | 0.17 |
ENSDART00000178638
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr19_+_43237078 | 0.16 |
ENSDART00000108775
|
ubp1
|
upstream binding protein 1 (LBP-1a) |
| chr3_-_47828170 | 0.16 |
|
|
|
| chr3_+_62051743 | 0.16 |
ENSDART00000168063
|
zgc:173575
|
zgc:173575 |
| chr25_-_1549206 | 0.16 |
|
|
|
| chr3_-_32409303 | 0.16 |
ENSDART00000151476
|
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr10_+_7744503 | 0.14 |
ENSDART00000099089
|
ggcx
|
gamma-glutamyl carboxylase |
| chr25_-_6262259 | 0.13 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr22_-_11618612 | 0.13 |
ENSDART00000109596
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr11_+_31061296 | 0.13 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr6_-_50588444 | 0.13 |
ENSDART00000151500
|
mtss1
|
metastasis suppressor 1 |
| chr10_-_44505679 | 0.12 |
|
|
|
| chr20_+_37963275 | 0.12 |
ENSDART00000153190
|
vash2
|
vasohibin 2 |
| chr4_+_295110 | 0.12 |
ENSDART00000067489
|
si:ch211-51e12.7
|
si:ch211-51e12.7 |
| chr11_-_18442169 | 0.12 |
ENSDART00000171183
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr17_+_31204729 | 0.11 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr5_+_25538112 | 0.11 |
ENSDART00000010041
|
dhfr
|
dihydrofolate reductase |
| chr19_-_34508527 | 0.11 |
ENSDART00000160495
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr11_-_15162471 | 0.10 |
ENSDART00000010684
|
rpn2
|
ribophorin II |
| chr20_-_40589999 | 0.10 |
ENSDART00000075070
|
hsf2
|
heat shock transcription factor 2 |
| chr9_-_43015354 | 0.10 |
|
|
|
| chr12_-_1018688 | 0.09 |
ENSDART00000054353
|
ENSDARG00000037357
|
ENSDARG00000037357 |
| chr4_+_75484112 | 0.09 |
ENSDART00000174393
|
si:dkey-240n22.8
|
si:dkey-240n22.8 |
| chr21_+_5597536 | 0.09 |
ENSDART00000161235
ENSDART00000170456 |
shroom3
|
shroom family member 3 |
| chr20_+_31312234 | 0.09 |
|
|
|
| chr12_-_36303384 | 0.09 |
ENSDART00000168538
|
oxld1
|
oxidoreductase-like domain containing 1 |
| chr5_+_61136704 | 0.08 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr7_-_73590802 | 0.08 |
ENSDART00000167855
|
FP236812.8
|
Histone H2B 1/2 |
| chr9_+_34412811 | 0.08 |
ENSDART00000028617
|
mpc2
|
mitochondrial pyruvate carrier 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.6 | 1.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 1.0 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.3 | 2.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.3 | 1.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.3 | 0.8 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 1.4 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.2 | 3.0 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.2 | 0.6 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.2 | 0.6 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 1.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.3 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 0.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.4 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 1.1 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.4 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.5 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.1 | 0.8 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 0.9 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.5 | GO:0090317 | negative regulation of intracellular protein transport(GO:0090317) |
| 0.1 | 1.6 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 1.0 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.4 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.4 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 2.6 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.6 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 0.5 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.5 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.1 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 1.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.2 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.3 | GO:0072015 | neural tube closure(GO:0001843) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.3 | 0.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.4 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.5 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 5.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.8 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.3 | 0.9 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 0.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 0.8 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.2 | 1.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 5.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.7 | GO:0015245 | long-chain fatty acid transporter activity(GO:0005324) fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.4 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.5 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 0.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.6 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.1 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.0 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 1.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 2.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |