Project
DANIO-CODE
Navigation
Downloads
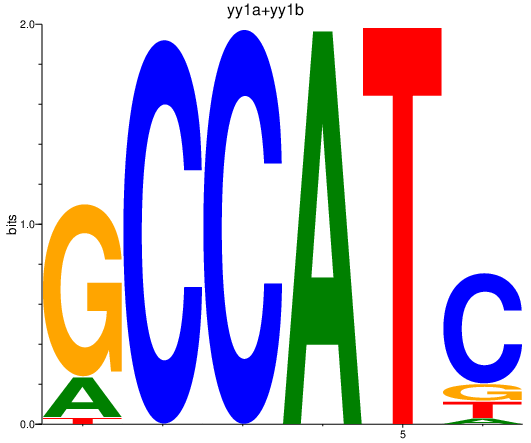
Results for yy1a+yy1b
Z-value: 1.63
Transcription factors associated with yy1a+yy1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
yy1b
|
ENSDARG00000027978 | YY1 transcription factor b |
|
yy1a
|
ENSDARG00000042796 | YY1 transcription factor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| yy1a | dr10_dc_chr17_-_30846176_30846335 | -0.56 | 2.4e-02 | Click! |
| yy1b | dr10_dc_chr20_-_54645287_54645324 | -0.39 | 1.4e-01 | Click! |
Activity profile of yy1a+yy1b motif
Sorted Z-values of yy1a+yy1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of yy1a+yy1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_18118467 | 2.64 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr14_-_35552296 | 2.50 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr12_+_22448972 | 1.97 |
|
|
|
| chr3_-_31942015 | 1.85 |
|
|
|
| chr14_-_41101660 | 1.71 |
ENSDART00000003170
|
mid1ip1l
|
MID1 interacting protein 1, like |
| chr21_-_13593659 | 1.65 |
ENSDART00000065817
|
pou5f3
|
POU domain, class 5, transcription factor 3 |
| chr12_-_16386055 | 1.60 |
ENSDART00000152750
|
pcgf5b
|
polycomb group ring finger 5b |
| chr18_+_284210 | 1.53 |
ENSDART00000158040
|
larp6
|
La ribonucleoprotein domain family, member 6 |
| chr2_+_10349452 | 1.46 |
ENSDART00000153678
|
pfn2l
|
profilin 2 like |
| chr5_+_24016742 | 1.45 |
ENSDART00000144560
|
dgcr8
|
DGCR8 microprocessor complex subunit |
| chr7_+_5843895 | 1.44 |
ENSDART00000115062
|
CU459186.4
|
Histone H3.2 |
| chr25_-_7490206 | 1.34 |
ENSDART00000163018
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr7_-_26226451 | 1.34 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr6_-_55342742 | 1.32 |
|
|
|
| chr10_-_25448769 | 1.32 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr14_+_31189527 | 1.31 |
ENSDART00000053026
|
fam122b
|
family with sequence similarity 122B |
| chr7_-_6309265 | 1.30 |
ENSDART00000172825
|
FP325123.5
|
Histone H3.2 |
| chr2_+_11423028 | 1.30 |
ENSDART00000144982
|
lhx8a
|
LIM homeobox 8a |
| chr10_-_26240772 | 1.30 |
ENSDART00000131394
|
fhdc3
|
FH2 domain containing 3 |
| chr14_+_1226767 | 1.29 |
ENSDART00000127477
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr5_+_23543441 | 1.27 |
ENSDART00000174856
|
ctdnep1a
|
CTD nuclear envelope phosphatase 1a |
| chr10_-_34058331 | 1.26 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr20_+_16740320 | 1.26 |
ENSDART00000152359
|
tmem30ab
|
transmembrane protein 30Ab |
| chr2_-_26941084 | 1.25 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr19_-_20819101 | 1.24 |
ENSDART00000137590
|
dazl
|
deleted in azoospermia-like |
| chr22_+_26840517 | 1.22 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr2_-_21512154 | 1.20 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr12_-_17370920 | 1.20 |
ENSDART00000130735
|
minpp1b
|
multiple inositol-polyphosphate phosphatase 1b |
| chr19_-_43359719 | 1.19 |
ENSDART00000086659
|
cyhr1
|
cysteine/histidine-rich 1 |
| chr6_-_7529384 | 1.19 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr21_+_3732973 | 1.19 |
ENSDART00000170653
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr19_-_30833687 | 1.18 |
ENSDART00000144121
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr16_+_48772036 | 1.16 |
ENSDART00000148709
|
brd2b
|
bromodomain containing 2b |
| chr15_+_30430282 | 1.14 |
ENSDART00000112784
|
lyrm9
|
LYR motif containing 9 |
| chr1_+_30840656 | 1.13 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr12_-_16385963 | 1.11 |
ENSDART00000152271
|
pcgf5b
|
polycomb group ring finger 5b |
| chr15_-_21733882 | 1.10 |
ENSDART00000156995
|
sorl1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr21_-_34227158 | 1.09 |
ENSDART00000169218
ENSDART00000064320 ENSDART00000172381 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr20_+_27341244 | 1.09 |
ENSDART00000123950
|
ENSDARG00000077377
|
ENSDARG00000077377 |
| chr14_+_14850200 | 1.08 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr20_+_23602537 | 1.07 |
|
|
|
| chr19_-_20819477 | 1.07 |
ENSDART00000151356
|
dazl
|
deleted in azoospermia-like |
| chr10_-_25448712 | 1.04 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr25_-_34597744 | 1.03 |
ENSDART00000153747
|
si:dkey-108k21.24
|
si:dkey-108k21.24 |
| chr7_+_44463158 | 1.03 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr5_-_13347964 | 1.03 |
ENSDART00000127109
|
figla
|
folliculogenesis specific bHLH transcription factor |
| chr19_+_41419851 | 1.01 |
ENSDART00000138555
ENSDART00000158587 ENSDART00000049842 |
casd1
|
CAS1 domain containing 1 |
| chr22_+_24291724 | 1.01 |
ENSDART00000168169
|
ccdc50
|
coiled-coil domain containing 50 |
| chr7_-_73897752 | 0.99 |
ENSDART00000164874
|
cldnd1a
|
claudin domain containing 1a |
| chr25_+_35799816 | 0.99 |
ENSDART00000103016
|
zgc:173552
|
zgc:173552 |
| chr15_+_37644266 | 0.99 |
ENSDART00000099456
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr4_-_15005050 | 0.98 |
ENSDART00000067048
|
klhdc10
|
kelch domain containing 10 |
| chr12_+_29017351 | 0.98 |
|
|
|
| chr7_+_69211965 | 0.98 |
ENSDART00000028064
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr25_-_9889107 | 0.97 |
ENSDART00000137407
|
AL929493.1
|
ENSDARG00000093575 |
| chr13_-_21541531 | 0.97 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr12_-_7201358 | 0.97 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr6_-_8125341 | 0.97 |
ENSDART00000139161
ENSDART00000140021 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr9_-_28444933 | 0.96 |
|
|
|
| chr3_-_15517844 | 0.96 |
ENSDART00000115022
|
zgc:66474
|
zgc:66474 |
| chr2_+_24982557 | 0.95 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr19_+_8693855 | 0.94 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr14_+_10351071 | 0.94 |
ENSDART00000061932
|
atrx
|
alpha thalassemia/mental retardation syndrome X-linked homolog (human) |
| chr19_+_8693695 | 0.94 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr24_-_23639458 | 0.93 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr2_-_24909432 | 0.93 |
ENSDART00000170283
|
crsp7
|
cofactor required for Sp1 transcriptional activation, subunit 7 |
| chr3_-_20989306 | 0.93 |
|
|
|
| chr23_-_34017934 | 0.93 |
ENSDART00000133223
|
ENSDARG00000070393
|
ENSDARG00000070393 |
| chr8_+_7625846 | 0.92 |
ENSDART00000175045
|
CABZ01078218.1
|
ENSDARG00000107577 |
| chr19_-_27093002 | 0.91 |
|
|
|
| chr12_+_48590478 | 0.91 |
|
|
|
| chr7_+_73595871 | 0.91 |
ENSDART00000169756
|
HIST1H2BA (1 of many)
|
Histone H2B 1/2 |
| chr24_-_30929990 | 0.91 |
|
|
|
| chr7_-_6309089 | 0.91 |
ENSDART00000172825
|
FP325123.5
|
Histone H3.2 |
| chr25_+_35799870 | 0.91 |
ENSDART00000103016
|
zgc:173552
|
zgc:173552 |
| chr1_+_157840 | 0.91 |
ENSDART00000152205
ENSDART00000160843 |
cul4a
|
cullin 4A |
| chr2_+_24982203 | 0.91 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr7_+_12675166 | 0.90 |
ENSDART00000027329
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr2_-_21512403 | 0.90 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr19_-_30833752 | 0.90 |
ENSDART00000144121
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr20_-_31840932 | 0.89 |
ENSDART00000137679
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr25_-_7546532 | 0.89 |
ENSDART00000131583
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr2_-_31703111 | 0.89 |
ENSDART00000126177
|
e2f5
|
E2F transcription factor 5 |
| chr2_+_24982611 | 0.88 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr18_+_15137751 | 0.88 |
ENSDART00000168639
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr9_-_32994156 | 0.87 |
ENSDART00000133382
|
zgc:112056
|
zgc:112056 |
| chr2_-_19585211 | 0.87 |
ENSDART00000169384
|
cdc20
|
cell division cycle 20 homolog |
| chr12_+_31668126 | 0.86 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr11_+_36117256 | 0.86 |
ENSDART00000141529
|
atxn7l2a
|
ataxin 7-like 2a |
| chr6_+_48188313 | 0.86 |
|
|
|
| chr10_+_2554651 | 0.86 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin-like 2 |
| chr19_-_7402246 | 0.85 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr7_+_5783560 | 0.85 |
ENSDART00000167099
|
CU459186.5
|
Histone H3.2 |
| chr10_-_20629538 | 0.84 |
|
|
|
| chr8_+_7625791 | 0.84 |
ENSDART00000175045
|
CABZ01078218.1
|
ENSDARG00000107577 |
| chr12_-_16386008 | 0.84 |
ENSDART00000152750
|
pcgf5b
|
polycomb group ring finger 5b |
| chr6_+_41506220 | 0.83 |
ENSDART00000136538
|
cish
|
cytokine inducible SH2-containing protein |
| chr7_-_73622585 | 0.83 |
ENSDART00000126541
|
zgc:173552
|
zgc:173552 |
| chr2_+_8314513 | 0.83 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr24_+_12693997 | 0.82 |
|
|
|
| chr4_-_72771281 | 0.82 |
ENSDART00000164550
ENSDART00000174320 |
RAB21
|
RAB21, member RAS oncogene family |
| chr2_-_31850133 | 0.82 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr6_+_21887963 | 0.82 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr22_+_24291633 | 0.81 |
ENSDART00000165618
|
ccdc50
|
coiled-coil domain containing 50 |
| KN149861v1_-_5543 | 0.81 |
|
|
|
| chr16_-_13723352 | 0.81 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| KN150065v1_-_622 | 0.80 |
|
|
|
| chr3_+_42395984 | 0.80 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr3_-_23466232 | 0.80 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr11_-_18078147 | 0.79 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr5_+_60823823 | 0.79 |
ENSDART00000023676
|
bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr16_-_24727689 | 0.79 |
ENSDART00000167121
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr18_+_22804237 | 0.79 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr8_-_39825675 | 0.79 |
ENSDART00000130686
|
unc119.1
|
unc-119 homolog 1 |
| chr9_+_24310158 | 0.79 |
ENSDART00000101565
|
MAIP1
|
matrix AAA peptidase interacting protein 1 |
| chr13_+_25313512 | 0.78 |
ENSDART00000131364
|
BX530079.1
|
ENSDARG00000094739 |
| chr1_-_30925005 | 0.78 |
ENSDART00000142296
|
dpcd
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr1_+_30840599 | 0.77 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr15_+_30430093 | 0.77 |
ENSDART00000112784
|
lyrm9
|
LYR motif containing 9 |
| chr21_+_21754830 | 0.77 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr3_-_15517767 | 0.77 |
ENSDART00000115022
|
zgc:66474
|
zgc:66474 |
| KN150589v1_-_5209 | 0.76 |
ENSDART00000157761
ENSDART00000157531 |
elovl7b
|
ELOVL fatty acid elongase 7b |
| chr14_-_30578373 | 0.76 |
ENSDART00000176631
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr13_-_22772707 | 0.76 |
ENSDART00000089133
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr10_-_25737220 | 0.76 |
ENSDART00000135058
|
sod1
|
superoxide dismutase 1, soluble |
| chr1_-_40671573 | 0.76 |
ENSDART00000074777
ENSDART00000158114 |
htt
|
huntingtin |
| chr18_+_18011759 | 0.76 |
ENSDART00000147797
|
ENSDARG00000011498
|
ENSDARG00000011498 |
| chr8_-_22265875 | 0.76 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr3_-_14545237 | 0.76 |
ENSDART00000133850
|
gadd45gip1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr2_+_24982471 | 0.76 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr20_+_39783565 | 0.76 |
ENSDART00000153142
|
hddc2
|
HD domain containing 2 |
| chr5_-_56641212 | 0.75 |
ENSDART00000050957
|
fer
|
fer (fps/fes related) tyrosine kinase |
| KN150563v1_-_10770 | 0.75 |
|
|
|
| chr24_-_42053682 | 0.75 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr23_-_24328931 | 0.75 |
|
|
|
| chr3_-_9755642 | 0.74 |
ENSDART00000168366
|
crebbpb
|
CREB binding protein b |
| chr12_-_41217465 | 0.74 |
ENSDART00000172175
|
ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr3_-_20989068 | 0.74 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr7_-_41601001 | 0.74 |
ENSDART00000174258
|
zgc:92818
|
zgc:92818 |
| chr17_-_31387062 | 0.74 |
|
|
|
| chr25_+_34629476 | 0.74 |
ENSDART00000156376
|
CR762436.2
|
Histone H3.2 |
| chr23_+_20586745 | 0.74 |
ENSDART00000157522
|
dpm1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr12_+_22449611 | 0.74 |
|
|
|
| chr24_+_14792755 | 0.74 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr6_+_41506350 | 0.73 |
ENSDART00000136538
|
cish
|
cytokine inducible SH2-containing protein |
| chr14_-_5100304 | 0.73 |
ENSDART00000168074
|
pcgf1
|
polycomb group ring finger 1 |
| chr8_-_49217824 | 0.73 |
|
|
|
| chr7_-_39467720 | 0.72 |
ENSDART00000052201
|
ccdc96
|
coiled-coil domain containing 96 |
| chr19_+_29871887 | 0.72 |
|
|
|
| chr2_-_23003710 | 0.72 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr7_+_30508612 | 0.72 |
ENSDART00000066806
|
mcee
|
methylmalonyl CoA epimerase |
| chr10_-_33307992 | 0.72 |
ENSDART00000169064
|
bcl7ba
|
B-cell CLL/lymphoma 7B, a |
| chr12_+_26943407 | 0.71 |
ENSDART00000153054
|
fbrs
|
fibrosin |
| chr16_-_26654610 | 0.71 |
ENSDART00000134448
|
l3mbtl1b
|
l(3)mbt-like 1b (Drosophila) |
| chr2_-_24909674 | 0.71 |
ENSDART00000170283
|
crsp7
|
cofactor required for Sp1 transcriptional activation, subunit 7 |
| chr17_+_19606608 | 0.71 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr7_-_6231474 | 0.71 |
ENSDART00000159524
|
CU457819.1
|
Histone H3.2 |
| chr24_+_17115897 | 0.70 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr2_-_31849916 | 0.70 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr8_+_25276000 | 0.70 |
ENSDART00000049793
|
gstm.1
|
glutathione S-transferase mu, tandem duplicate 1 |
| chr7_+_34923319 | 0.70 |
|
|
|
| KN149861v1_-_5655 | 0.69 |
|
|
|
| chr24_+_28304276 | 0.69 |
ENSDART00000018095
|
sh3glb1a
|
SH3-domain GRB2-like endophilin B1a |
| chr21_-_43462380 | 0.69 |
ENSDART00000085039
|
stk26
|
serine/threonine protein kinase 26 |
| chr11_-_25223594 | 0.69 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr15_+_14656797 | 0.69 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr2_+_24982268 | 0.69 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr20_-_7079412 | 0.69 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr12_-_35467285 | 0.69 |
ENSDART00000160336
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr16_-_39291647 | 0.68 |
ENSDART00000171342
|
tmem42a
|
transmembrane protein 42a |
| chr8_+_17107665 | 0.68 |
ENSDART00000158707
ENSDART00000061758 |
mier3b
|
mesoderm induction early response 1, family member 3 b |
| chr24_-_10756995 | 0.68 |
ENSDART00000145593
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr25_+_34629404 | 0.68 |
ENSDART00000156376
|
CR762436.2
|
Histone H3.2 |
| chr2_-_21512285 | 0.68 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr6_-_3763940 | 0.68 |
ENSDART00000171804
|
tlk1b
|
tousled-like kinase 1b |
| chr19_-_19452999 | 0.67 |
ENSDART00000163359
ENSDART00000167951 |
dync1li1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr5_+_60823598 | 0.67 |
ENSDART00000023676
|
bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr8_-_4270732 | 0.67 |
ENSDART00000134378
|
cux2b
|
cut-like homeobox 2b |
| chr3_+_11985816 | 0.67 |
ENSDART00000081367
|
dnaja3a
|
DnaJ (Hsp40) homolog, subfamily A, member 3A |
| chr16_-_42236126 | 0.67 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr19_+_32178335 | 0.67 |
|
|
|
| chr3_+_1451017 | 0.67 |
ENSDART00000161945
|
rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr5_+_3567992 | 0.66 |
ENSDART00000129329
|
rpain
|
RPA interacting protein |
| chr10_-_33307845 | 0.66 |
ENSDART00000169064
|
bcl7ba
|
B-cell CLL/lymphoma 7B, a |
| chr8_-_22266041 | 0.66 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr2_-_19705537 | 0.66 |
ENSDART00000168627
|
zfyve9a
|
zinc finger, FYVE domain containing 9a |
| chr10_+_22885419 | 0.65 |
ENSDART00000079437
|
ap4m1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr19_-_7402329 | 0.65 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr22_+_22413803 | 0.65 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr2_-_23003738 | 0.65 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr13_-_30523358 | 0.65 |
|
|
|
| chr17_+_43749940 | 0.65 |
ENSDART00000133317
ENSDART00000140316 ENSDART00000142929 ENSDART00000133874 |
zgc:66313
|
zgc:66313 |
| chr24_+_17116228 | 0.65 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr2_-_26948396 | 0.65 |
ENSDART00000143085
|
yipf1
|
Yip1 domain family, member 1 |
| chr6_-_12687107 | 0.64 |
ENSDART00000155360
|
CU639412.1
|
ENSDARG00000096929 |
| chr20_-_42344217 | 0.64 |
ENSDART00000034054
|
nus1
|
NUS1 dehydrodolichyl diphosphate synthase subunit |
| chr7_-_43518699 | 0.64 |
|
|
|
| chr19_-_18689437 | 0.64 |
ENSDART00000016135
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr16_+_13795468 | 0.64 |
ENSDART00000142428
|
josd2
|
Josephin domain containing 2 |
| chr23_-_33783345 | 0.64 |
ENSDART00000143333
|
pou6f1
|
POU class 6 homeobox 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.0 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.6 | 3.2 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.6 | 4.2 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.4 | 2.1 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.4 | 1.2 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.4 | 1.2 | GO:0021990 | neural plate formation(GO:0021990) brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) anterior neural plate formation(GO:0090017) |
| 0.3 | 2.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.3 | 1.5 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.3 | 1.4 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.3 | 0.8 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.3 | 1.9 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.3 | 1.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.3 | 1.6 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 0.7 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.2 | 0.7 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.2 | 1.2 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.2 | 0.7 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.2 | 0.9 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.2 | 0.7 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 0.9 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 1.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 2.1 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.2 | 0.6 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.2 | 0.6 | GO:0070657 | mechanosensory epithelium regeneration(GO:0070655) mechanoreceptor differentiation involved in mechanosensory epithelium regeneration(GO:0070656) neuromast regeneration(GO:0070657) neuromast hair cell differentiation involved in neuromast regeneration(GO:0070658) |
| 0.2 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 1.4 | GO:0045453 | bone resorption(GO:0045453) |
| 0.2 | 1.0 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.2 | 0.2 | GO:0039531 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) cellular response to virus(GO:0098586) |
| 0.2 | 1.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 0.6 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.2 | 0.7 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 0.9 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 1.3 | GO:0008585 | female gonad development(GO:0008585) |
| 0.2 | 0.7 | GO:0061698 | regulation of ketone biosynthetic process(GO:0010566) protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) |
| 0.2 | 0.7 | GO:0036088 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.2 | 1.6 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 1.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 1.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 0.9 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.2 | 0.5 | GO:1902633 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process(GO:1902633) 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process(GO:1902635) |
| 0.2 | 0.5 | GO:0033238 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.2 | 0.6 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.2 | 1.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.6 | GO:0038066 | p38MAPK cascade(GO:0038066) positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.6 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.1 | 0.7 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.9 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 2.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.4 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.5 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.9 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 0.5 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.5 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.6 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 0.8 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.9 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 1.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.4 | GO:0043393 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.1 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 1.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.8 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.0 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.4 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.1 | 0.2 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.6 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.4 | GO:0006226 | dUMP biosynthetic process(GO:0006226) dUMP metabolic process(GO:0046078) |
| 0.1 | 1.3 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.6 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 1.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.0 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.3 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) galactolipid catabolic process(GO:0019376) |
| 0.1 | 1.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.8 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 0.4 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.1 | 0.3 | GO:1900144 | epithelial cell morphogenesis involved in gastrulation(GO:0003381) BMP secretion(GO:0038055) positive regulation of BMP secretion(GO:1900144) regulation of BMP secretion(GO:2001284) |
| 0.1 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 1.0 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 0.4 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.4 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 2.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.4 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.1 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.8 | GO:2000758 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 | 0.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 1.5 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.2 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.1 | 0.4 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 1.9 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.0 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.1 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.6 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.1 | 0.4 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 1.1 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 0.3 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.5 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.2 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 0.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.7 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.2 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 1.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.2 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.6 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.4 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 3.7 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.1 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 1.7 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 2.7 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.1 | 0.2 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) positive regulation of protein deubiquitination(GO:1903003) positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 1.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.6 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 1.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.7 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.7 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.2 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.5 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.5 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.5 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 1.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.8 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.0 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.5 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 5.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.2 | GO:0031058 | positive regulation of histone modification(GO:0031058) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.5 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.7 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.0 | 0.4 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.7 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.3 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.5 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.3 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.0 | 0.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.5 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0010482 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.3 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.3 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.3 | GO:0022406 | membrane docking(GO:0022406) |
| 0.0 | 0.2 | GO:0071174 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.3 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.2 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.1 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.0 | GO:0060911 | cardiac cell fate commitment(GO:0060911) |
| 0.0 | 0.0 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.4 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0051654 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.3 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.4 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.2 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 1.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:0034121 | regulation of toll-like receptor signaling pathway(GO:0034121) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.4 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.2 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.7 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.4 | GO:0070304 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.0 | 0.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0043650 | dicarboxylic acid biosynthetic process(GO:0043650) |
| 0.0 | 0.2 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.3 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.5 | 1.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.4 | 2.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.4 | 9.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 4.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 1.0 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.3 | 1.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.3 | 1.0 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.3 | 1.9 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.7 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 1.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 0.9 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.2 | 0.8 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.2 | 0.5 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 1.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 2.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 0.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 2.5 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.1 | 0.8 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.4 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.1 | 1.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.3 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 2.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.7 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.6 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.1 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 6.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.7 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 1.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 3.8 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 0.2 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.2 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 2.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 2.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.6 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.4 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.4 | 1.3 | GO:0031730 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 4.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 1.2 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.3 | 1.0 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.3 | 1.3 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.3 | 0.9 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.3 | 0.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.3 | 1.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 2.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 1.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 0.7 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.2 | 0.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 1.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 0.6 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 0.2 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 0.6 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 0.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 2.0 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 0.6 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 0.7 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 0.7 | GO:0036055 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.2 | 1.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 0.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 4.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 1.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 0.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 2.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 0.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.5 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 0.7 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.1 | 2.5 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 1.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.8 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 1.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.8 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.3 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.3 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.6 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.5 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.4 | GO:0001006 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 0.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.9 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.2 | GO:0035515 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.5 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 6.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.4 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.7 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 2.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0071424 | rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) |
| 0.0 | 0.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.5 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.0 | 6.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 4.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.7 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0008465 | glycerate dehydrogenase activity(GO:0008465) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 2.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.3 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 5.5 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.8 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.7 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.3 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0008665 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.4 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0071949 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) FAD binding(GO:0071949) |
| 0.0 | 1.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.2 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 2.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.2 | 0.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.1 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.1 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.9 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.5 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.2 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |