Project
DANIO-CODE
Navigation
Downloads
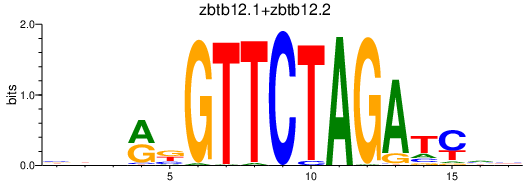
Results for zbtb12.1+zbtb12.2
Z-value: 0.70
Transcription factors associated with zbtb12.1+zbtb12.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb12.2
|
ENSDARG00000070658 | zinc finger and BTB domain containing 12, tandem duplicate 2 |
|
zbtb12.1
|
ENSDARG00000103949 | zinc finger and BTB domain containing 12, tandem duplicate 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb12.2 | dr10_dc_chr19_-_27379989_27380198 | -0.50 | 4.6e-02 | Click! |
Activity profile of zbtb12.1+zbtb12.2 motif
Sorted Z-values of zbtb12.1+zbtb12.2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb12.1+zbtb12.2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_17443607 | 2.06 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr15_+_38397772 | 1.90 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr19_-_27995593 | 1.76 |
ENSDART00000142263
|
si:dkeyp-46h3.3
|
si:dkeyp-46h3.3 |
| chr2_-_17443642 | 1.74 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr24_-_7928026 | 1.42 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| KN150339v1_-_39357 | 1.32 |
ENSDART00000169638
|
wu:fa19b12
|
wu:fa19b12 |
| chr8_-_38284959 | 1.23 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr21_+_20350152 | 1.20 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr14_-_6220150 | 1.18 |
ENSDART00000125058
|
nipsnap3a
|
nipsnap homolog 3A (C. elegans) |
| chr10_-_25383262 | 1.14 |
ENSDART00000166348
|
cct8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr19_+_21209328 | 1.13 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr12_+_9811557 | 1.10 |
ENSDART00000110458
|
fam117ab
|
family with sequence similarity 117, member Ab |
| chr21_-_28700679 | 1.00 |
ENSDART00000128237
ENSDART00000124826 |
nrg2a
|
neuregulin 2a |
| chr6_-_1613740 | 0.99 |
ENSDART00000156305
|
trim107
|
tripartite motif containing 107 |
| chr5_-_66145078 | 0.94 |
ENSDART00000041441
|
stip1
|
stress-induced phosphoprotein 1 |
| chr13_-_7904111 | 0.92 |
|
|
|
| chr14_+_47368995 | 0.91 |
ENSDART00000060577
|
tmem33
|
transmembrane protein 33 |
| chr19_+_27056002 | 0.89 |
ENSDART00000175951
|
ints3
|
integrator complex subunit 3 |
| chr22_-_9832436 | 0.87 |
|
|
|
| chr22_+_1283662 | 0.84 |
ENSDART00000124161
|
si:ch73-138e16.5
|
si:ch73-138e16.5 |
| chr20_-_5992604 | 0.83 |
ENSDART00000170468
|
cep128
|
centrosomal protein 128 |
| chr7_-_59851075 | 0.77 |
ENSDART00000128363
|
cct7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr17_-_12095843 | 0.73 |
ENSDART00000155545
|
ahctf1
|
AT hook containing transcription factor 1 |
| chr6_-_1613915 | 0.73 |
ENSDART00000112118
|
trim107
|
tripartite motif containing 107 |
| chr17_-_12095723 | 0.72 |
ENSDART00000115321
|
ahctf1
|
AT hook containing transcription factor 1 |
| chr19_+_27055869 | 0.72 |
ENSDART00000067793
ENSDART00000124755 |
ints3
|
integrator complex subunit 3 |
| chr17_+_49540985 | 0.70 |
|
|
|
| chr8_+_13757024 | 0.70 |
|
|
|
| chr5_-_45244910 | 0.69 |
|
|
|
| chr16_-_26505960 | 0.61 |
ENSDART00000144462
|
BX571757.1
|
ENSDARG00000088497 |
| chr8_+_13757180 | 0.61 |
|
|
|
| chr2_+_52225622 | 0.61 |
ENSDART00000170353
|
ENSDARG00000103108
|
ENSDARG00000103108 |
| chr14_+_47368951 | 0.60 |
ENSDART00000060577
|
tmem33
|
transmembrane protein 33 |
| chr15_+_38397897 | 0.59 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr22_-_9832520 | 0.59 |
|
|
|
| chr17_-_52239876 | 0.57 |
|
|
|
| chr21_-_13575544 | 0.57 |
ENSDART00000146717
|
clic3
|
chloride intracellular channel 3 |
| chr21_-_28700622 | 0.57 |
ENSDART00000098696
|
nrg2a
|
neuregulin 2a |
| chr6_-_1613994 | 0.55 |
ENSDART00000112118
|
trim107
|
tripartite motif containing 107 |
| chr4_-_4603205 | 0.54 |
ENSDART00000130601
|
CABZ01020840.1
|
ENSDARG00000090401 |
| chr10_+_29963102 | 0.53 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr6_-_2768437 | 0.50 |
|
|
|
| chr20_-_29630048 | 0.49 |
ENSDART00000049224
|
taf1b
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, B |
| chr18_+_8444064 | 0.49 |
ENSDART00000171974
|
prpf18
|
PRP18 pre-mRNA processing factor 18 homolog (yeast) |
| chr16_-_10946422 | 0.48 |
ENSDART00000036891
|
rabac1
|
Rab acceptor 1 (prenylated) |
| chr16_-_32865452 | 0.47 |
|
|
|
| chr7_+_28753073 | 0.46 |
ENSDART00000076425
|
cog4
|
component of oligomeric golgi complex 4 |
| chr17_-_35290338 | 0.46 |
ENSDART00000005784
|
itgb1bp1
|
integrin beta 1 binding protein 1 |
| chr3_-_36235222 | 0.43 |
ENSDART00000028883
|
gna13b
|
guanine nucleotide binding protein (G protein), alpha 13b |
| chr10_-_22176478 | 0.43 |
|
|
|
| chr21_-_43176006 | 0.38 |
ENSDART00000148630
|
hspa4a
|
heat shock protein 4a |
| chr19_+_9066225 | 0.38 |
ENSDART00000018973
|
scamp3
|
secretory carrier membrane protein 3 |
| chr19_+_21208954 | 0.37 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr8_-_38285034 | 0.36 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr16_-_42225498 | 0.36 |
ENSDART00000084715
|
alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr3_+_42667814 | 0.36 |
ENSDART00000172425
|
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr7_+_28753102 | 0.33 |
ENSDART00000076425
|
cog4
|
component of oligomeric golgi complex 4 |
| chr24_-_7928168 | 0.32 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| chr20_+_38555185 | 0.29 |
ENSDART00000020153
|
adck3
|
aarF domain containing kinase 3 |
| chr8_+_44629016 | 0.29 |
|
|
|
| chr16_-_48488178 | 0.27 |
ENSDART00000005927
|
rad21a
|
RAD21 cohesin complex component a |
| chr10_-_25383076 | 0.24 |
ENSDART00000009477
|
cct8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr2_+_52225552 | 0.23 |
ENSDART00000170353
|
ENSDARG00000103108
|
ENSDARG00000103108 |
| KN150650v1_+_861 | 0.22 |
|
|
|
| chr17_-_27402265 | 0.22 |
ENSDART00000127043
|
ythdf2
|
YTH N(6)-methyladenosine RNA binding protein 2 |
| chr12_-_17013870 | 0.22 |
|
|
|
| chr9_-_45096523 | 0.22 |
ENSDART00000110697
|
dnajc10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr10_-_15090551 | 0.21 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr1_-_51086075 | 0.20 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| KN150650v1_+_738 | 0.18 |
|
|
|
| chr7_-_59851104 | 0.15 |
ENSDART00000128363
|
cct7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr18_-_21921898 | 0.14 |
ENSDART00000132381
|
edc4
|
enhancer of mRNA decapping 4 |
| chr12_-_31619559 | 0.13 |
|
|
|
| chr3_+_40265548 | 0.09 |
ENSDART00000103486
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr7_-_56465497 | 0.08 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr19_+_43526327 | 0.07 |
|
|
|
| chr6_-_20308805 | 0.06 |
ENSDART00000083038
|
nprl2
|
NPR2-like, GATOR1 complex subunit |
| chr6_+_34406796 | 0.06 |
|
|
|
| chr16_-_10946309 | 0.05 |
ENSDART00000049148
|
rabac1
|
Rab acceptor 1 (prenylated) |
| chr6_+_40525610 | 0.05 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr5_-_42350607 | 0.02 |
ENSDART00000149868
|
grsf1
|
G-rich RNA sequence binding factor 1 |
| chr19_+_43526408 | 0.02 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.3 | 3.8 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.3 | 1.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.3 | 1.5 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.3 | 0.8 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.3 | 1.6 | GO:0098773 | ventricular trabecula myocardium morphogenesis(GO:0003222) skin epidermis development(GO:0098773) |
| 0.1 | 0.5 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.5 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.4 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.1 | 1.6 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 0.5 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.1 | 0.2 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.1 | 1.7 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.3 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 2.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 0.5 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 0.5 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 1.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.3 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 3.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 1.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.4 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 2.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 1.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 2.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |