Project
DANIO-CODE
Navigation
Downloads
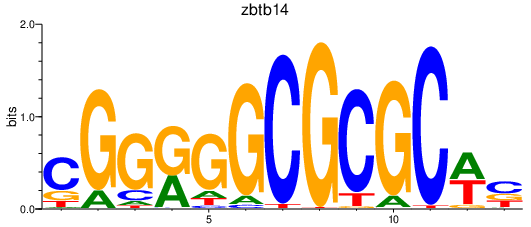
Results for zbtb14
Z-value: 2.26
Transcription factors associated with zbtb14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb14
|
ENSDARG00000098273 | zinc finger and BTB domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb14 | dr10_dc_chr24_-_42053682_42053815 | -0.32 | 2.3e-01 | Click! |
Activity profile of zbtb14 motif
Sorted Z-values of zbtb14 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb14
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_32261533 | 7.60 |
ENSDART00000003745
|
vim
|
vimentin |
| chr1_+_53286155 | 5.77 |
ENSDART00000139625
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr4_+_5733160 | 5.67 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr21_+_105327 | 5.14 |
ENSDART00000007672
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr18_+_62936 | 4.90 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr23_-_20332076 | 4.73 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr4_-_73485204 | 4.52 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr11_+_15796506 | 4.37 |
ENSDART00000081098
|
draxin
|
dorsal inhibitory axon guidance protein |
| chr7_+_67227810 | 4.22 |
ENSDART00000163840
|
gcshb
|
glycine cleavage system protein H (aminomethyl carrier), b |
| chr2_-_58737903 | 4.05 |
|
|
|
| chr1_-_55554518 | 3.90 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr10_-_27261937 | 3.81 |
|
|
|
| chr2_+_42874975 | 3.66 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr4_+_22959458 | 3.61 |
ENSDART00000036531
|
gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr20_-_26632676 | 3.61 |
ENSDART00000131994
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr12_-_7572970 | 3.55 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr10_-_13814731 | 3.52 |
ENSDART00000145103
|
cntfr
|
ciliary neurotrophic factor receptor |
| chr19_+_3713027 | 3.49 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr4_+_9027862 | 3.45 |
ENSDART00000102893
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr22_-_13018196 | 3.30 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr23_-_39956151 | 3.28 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr23_+_45876483 | 3.19 |
ENSDART00000169521
ENSDART00000162915 |
dclk2b
|
doublecortin-like kinase 2b |
| chr15_+_37039861 | 3.09 |
ENSDART00000172664
|
kirrel3l
|
kin of IRRE like 3 like |
| chr4_+_10065500 | 3.07 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr19_+_26065401 | 3.02 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr1_-_37990863 | 3.02 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr20_-_35343242 | 2.96 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr22_-_3661536 | 2.95 |
ENSDART00000153634
|
FP565432.1
|
ENSDARG00000097145 |
| chr23_+_45020761 | 2.89 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr14_-_25279835 | 2.88 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr10_-_37131530 | 2.88 |
ENSDART00000132023
|
myo18aa
|
myosin XVIIIAa |
| chr2_+_26523457 | 2.84 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr16_+_3090170 | 2.83 |
ENSDART00000110395
|
limd1a
|
LIM domains containing 1a |
| chr17_-_756401 | 2.82 |
ENSDART00000157622
|
zbtb42
|
zinc finger and BTB domain containing 42 |
| chr14_+_14759314 | 2.79 |
ENSDART00000167075
|
nkx1.2lb
|
NK1 transcription factor related 2-like,b |
| chr5_+_49103778 | 2.79 |
ENSDART00000155405
|
CR751602.2
|
ENSDARG00000098045 |
| chr2_-_30340646 | 2.79 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr9_-_14533551 | 2.77 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr4_-_73485028 | 2.74 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr8_-_387126 | 2.73 |
|
|
|
| chr5_+_62987426 | 2.70 |
ENSDART00000178937
|
dnm1b
|
dynamin 1b |
| chr13_-_5440923 | 2.69 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr6_+_56157608 | 2.68 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr7_-_14135850 | 2.62 |
ENSDART00000161442
|
CU467822.1
|
ENSDARG00000101584 |
| chr3_+_17594555 | 2.55 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr20_+_54512601 | 2.52 |
ENSDART00000169386
|
faua
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed a |
| chr3_+_13114740 | 2.50 |
ENSDART00000162724
|
ENSDARG00000074231
|
ENSDARG00000074231 |
| chr20_-_40553998 | 2.50 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr24_-_38928988 | 2.49 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr23_+_45876447 | 2.47 |
ENSDART00000169521
ENSDART00000162915 |
dclk2b
|
doublecortin-like kinase 2b |
| chr12_-_9400344 | 2.46 |
ENSDART00000003932
|
erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr11_-_1108157 | 2.46 |
|
|
|
| chr18_+_45680282 | 2.45 |
ENSDART00000109948
|
qser1
|
glutamine and serine rich 1 |
| chr5_-_43896815 | 2.45 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr17_+_3842688 | 2.43 |
ENSDART00000170822
|
tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr22_-_21289850 | 2.42 |
ENSDART00000135388
|
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr17_+_53224434 | 2.41 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr5_-_43896757 | 2.40 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr3_-_21149752 | 2.39 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr22_+_223797 | 2.36 |
|
|
|
| chr1_-_22144014 | 2.36 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr5_-_15774816 | 2.35 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr19_-_5338129 | 2.34 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr25_-_35760878 | 2.32 |
ENSDART00000121786
|
hist1h4l
|
histone 1, H4, like |
| chr23_+_2193582 | 2.31 |
ENSDART00000106336
|
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr13_+_1741432 | 2.31 |
ENSDART00000161162
|
bmp5
|
bone morphogenetic protein 5 |
| chr25_-_18044103 | 2.31 |
ENSDART00000113581
|
kitlga
|
kit ligand a |
| chr13_-_31339870 | 2.30 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| KN150001v1_+_14939 | 2.29 |
|
|
|
| chr4_+_3970874 | 2.27 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr25_-_20283541 | 2.26 |
ENSDART00000142665
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr13_+_1741474 | 2.26 |
ENSDART00000161162
|
bmp5
|
bone morphogenetic protein 5 |
| chr25_+_34997593 | 2.25 |
ENSDART00000058443
|
fibina
|
fin bud initiation factor a |
| chr24_-_35811390 | 2.25 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_+_49683715 | 2.23 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr16_+_28819826 | 2.20 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr23_-_44460486 | 2.16 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr16_+_32605735 | 2.16 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr4_+_14972481 | 2.15 |
ENSDART00000005985
|
smo
|
smoothened, frizzled class receptor |
| chr8_-_50899451 | 2.13 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long-chain family member 2 |
| chr14_-_46219646 | 2.13 |
ENSDART00000110191
|
shisa3
|
shisa family member 3 |
| chr12_-_48493654 | 2.09 |
ENSDART00000162603
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr4_+_14972708 | 2.08 |
ENSDART00000005985
|
smo
|
smoothened, frizzled class receptor |
| chr11_-_37322205 | 2.08 |
ENSDART00000172989
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr19_+_1626598 | 2.07 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr18_-_11426 | 2.05 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr17_+_9150326 | 2.05 |
|
|
|
| chr9_-_5618609 | 2.05 |
ENSDART00000127219
|
FAM155A
|
family with sequence similarity 155 member A |
| chr6_+_12619062 | 2.04 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr19_+_41488385 | 2.03 |
ENSDART00000138687
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr20_+_4078243 | 2.03 |
ENSDART00000124197
|
CU929133.1
|
ENSDARG00000088444 |
| chr7_-_37587227 | 2.03 |
ENSDART00000173552
|
adcy7
|
adenylate cyclase 7 |
| chr17_+_38619041 | 2.01 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| KN150349v1_-_13313 | 2.01 |
|
|
|
| chr9_-_695593 | 1.98 |
ENSDART00000115030
|
dip2a
|
disco-interacting protein 2 homolog A |
| chr3_-_45779817 | 1.96 |
ENSDART00000164361
|
gcgra
|
glucagon receptor a |
| chr2_-_53156364 | 1.96 |
|
|
|
| chr25_-_22541525 | 1.95 |
ENSDART00000073583
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr20_-_35343057 | 1.94 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr16_+_23172295 | 1.93 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr24_-_28166614 | 1.92 |
ENSDART00000167861
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr1_+_19942293 | 1.90 |
ENSDART00000148518
|
spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr5_-_20527105 | 1.89 |
ENSDART00000133461
|
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr21_+_142265 | 1.89 |
|
|
|
| chr12_-_1931281 | 1.88 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr3_-_1435654 | 1.88 |
ENSDART00000089646
ENSDART00000158110 |
fam234b
|
family with sequence similarity 234, member B |
| chr2_+_46179589 | 1.88 |
ENSDART00000125971
|
gpc1b
|
glypican 1b |
| chr2_+_49358871 | 1.87 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr13_+_35619748 | 1.84 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr19_-_22904687 | 1.83 |
ENSDART00000141503
|
pleca
|
plectin a |
| chr7_-_64637419 | 1.79 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr3_-_21217799 | 1.79 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr25_+_34515221 | 1.78 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr1_-_35419335 | 1.76 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr11_+_41246824 | 1.76 |
ENSDART00000161605
|
iffo2b
|
intermediate filament family orphan 2b |
| chr23_-_1124499 | 1.76 |
|
|
|
| chr20_+_1724609 | 1.75 |
|
|
|
| chr24_+_4946079 | 1.73 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr11_-_36503807 | 1.71 |
ENSDART00000002055
|
cacna2d3
|
calcium channel, voltage dependent, alpha2/delta subunit 3 |
| chr11_-_33925792 | 1.71 |
ENSDART00000172978
|
atp13a3
|
ATPase type 13A3 |
| chr12_-_954375 | 1.71 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr24_+_10273081 | 1.70 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr24_+_5811389 | 1.66 |
ENSDART00000139191
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr9_+_8951400 | 1.66 |
ENSDART00000102562
|
ankrd10b
|
ankyrin repeat domain 10b |
| KN149883v1_-_5164 | 1.66 |
ENSDART00000158506
ENSDART00000172360 |
NDUFA13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr18_-_25190648 | 1.65 |
ENSDART00000175178
|
slco3a1
|
solute carrier organic anion transporter family, member 3A1 |
| chr20_-_1293519 | 1.64 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr8_-_4903896 | 1.64 |
ENSDART00000004986
|
igsf9b
|
immunoglobulin superfamily, member 9b |
| chr8_+_7237516 | 1.62 |
|
|
|
| chr1_-_50215233 | 1.62 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr4_-_5755663 | 1.62 |
ENSDART00000113864
|
faxca
|
failed axon connections homolog a |
| chr5_+_68943914 | 1.61 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr19_-_35512932 | 1.61 |
|
|
|
| chr14_+_51653588 | 1.59 |
ENSDART00000157833
|
igfbp7
|
insulin-like growth factor binding protein 7 |
| chr20_+_4222097 | 1.59 |
ENSDART00000097543
|
im:7142702
|
im:7142702 |
| chr25_-_12107407 | 1.59 |
ENSDART00000159800
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr20_-_8431338 | 1.57 |
ENSDART00000145841
ENSDART00000143936 ENSDART00000083906 ENSDART00000167102 ENSDART00000083908 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr25_-_20283460 | 1.54 |
ENSDART00000067454
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr20_+_26993709 | 1.54 |
ENSDART00000133293
|
ftr97
|
finTRIM family, member 97 |
| chr18_+_44802349 | 1.54 |
ENSDART00000139526
|
fam118b
|
family with sequence similarity 118, member B |
| chr20_+_51034244 | 1.53 |
|
|
|
| chr19_-_35513216 | 1.52 |
|
|
|
| chr17_-_6191539 | 1.52 |
ENSDART00000081707
|
ENSDARG00000058775
|
ENSDARG00000058775 |
| chr20_-_8431368 | 1.52 |
ENSDART00000145841
ENSDART00000143936 ENSDART00000083906 ENSDART00000167102 ENSDART00000083908 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr24_+_16249188 | 1.50 |
ENSDART00000164516
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr12_+_26530620 | 1.50 |
ENSDART00000046959
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr6_+_4379989 | 1.49 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr20_-_40217185 | 1.49 |
|
|
|
| KN149842v1_+_13298 | 1.49 |
ENSDART00000166541
|
CABZ01101812.1
|
ENSDARG00000098297 |
| chr20_-_20412830 | 1.49 |
ENSDART00000114779
|
ENSDARG00000079369
|
ENSDARG00000079369 |
| chr10_+_45499009 | 1.49 |
ENSDART00000166085
|
ppiab
|
peptidylprolyl isomerase Ab (cyclophilin A) |
| chr14_-_49600662 | 1.47 |
ENSDART00000161147
|
si:ch211-154c21.1
|
si:ch211-154c21.1 |
| chr7_+_39353662 | 1.46 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr23_-_19177660 | 1.46 |
ENSDART00000016901
|
pard6b
|
par-6 partitioning defective 6 homolog beta (C. elegans) |
| chr11_-_33697442 | 1.46 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr12_+_1370058 | 1.45 |
ENSDART00000026303
|
rasd1
|
RAS, dexamethasone-induced 1 |
| chr10_-_21096635 | 1.44 |
ENSDART00000091004
|
pcdh1a
|
protocadherin 1a |
| chr12_-_37124892 | 1.43 |
ENSDART00000146142
|
pmp22b
|
peripheral myelin protein 22b |
| chr7_-_49322226 | 1.43 |
ENSDART00000174161
|
brsk2b
|
BR serine/threonine kinase 2b |
| chr4_-_9172622 | 1.43 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr20_-_54942216 | 1.42 |
|
|
|
| chr25_+_34556009 | 1.41 |
ENSDART00000111706
|
si:dkey-108k21.14
|
si:dkey-108k21.14 |
| chr1_-_7744605 | 1.41 |
ENSDART00000033917
|
syngr3b
|
synaptogyrin 3b |
| chr19_+_47846675 | 1.41 |
ENSDART00000041114
|
psmb2
|
proteasome subunit beta 2 |
| chr11_-_10844854 | 1.40 |
ENSDART00000040180
|
tbr1a
|
T-box, brain, 1a |
| chr4_-_73485124 | 1.39 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr23_-_7740845 | 1.39 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr13_+_29924636 | 1.38 |
ENSDART00000110061
|
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr8_-_53584884 | 1.38 |
|
|
|
| chr9_-_47041677 | 1.37 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr1_-_47080507 | 1.37 |
ENSDART00000101079
|
neurl1aa
|
neuralized E3 ubiquitin protein ligase 1Aa |
| chr13_-_36008571 | 1.37 |
ENSDART00000139593
|
pcnx
|
pecanex homolog (Drosophila) |
| chr6_+_56163589 | 1.36 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr22_-_6977038 | 1.36 |
ENSDART00000133143
ENSDART00000146813 |
gpd1b
|
glycerol-3-phosphate dehydrogenase 1b |
| chr14_+_14759351 | 1.35 |
ENSDART00000167075
|
nkx1.2lb
|
NK1 transcription factor related 2-like,b |
| chr20_+_48950833 | 1.34 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr4_-_25080470 | 1.33 |
ENSDART00000179640
|
gata3
|
GATA binding protein 3 |
| chr12_-_45714172 | 1.33 |
ENSDART00000149044
|
pax2b
|
paired box 2b |
| KN150487v1_+_38777 | 1.33 |
|
|
|
| chr12_-_1545172 | 1.33 |
|
|
|
| chr23_+_36648211 | 1.33 |
|
|
|
| chr12_-_28248133 | 1.33 |
ENSDART00000016283
|
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr14_-_6624878 | 1.32 |
ENSDART00000166439
|
si:ch211-266k2.1
|
si:ch211-266k2.1 |
| chr22_-_4622401 | 1.32 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr7_+_73371895 | 1.32 |
ENSDART00000110544
|
znf219
|
zinc finger protein 219 |
| chr19_+_19839981 | 1.31 |
ENSDART00000113580
|
ENSDARG00000074216
|
ENSDARG00000074216 |
| chr4_-_22751641 | 1.31 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr25_-_211879 | 1.31 |
ENSDART00000021614
|
srpk2
|
SRSF protein kinase 2 |
| chr7_+_32451041 | 1.30 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr3_-_42649955 | 1.29 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr16_-_204751 | 1.29 |
|
|
|
| chr13_+_51769487 | 1.28 |
ENSDART00000169928
ENSDART00000160698 |
klhdc3
|
kelch domain containing 3 |
| chr1_-_37990935 | 1.28 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr21_-_661222 | 1.28 |
|
|
|
| chr7_-_11812634 | 1.28 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr15_+_46813538 | 1.28 |
|
|
|
| chr10_-_27262050 | 1.27 |
|
|
|
| chr11_-_45008060 | 1.26 |
ENSDART00000166691
|
trappc10
|
trafficking protein particle complex 10 |
| chr4_-_12008472 | 1.26 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr23_-_104449 | 1.24 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.8 | 4.2 | GO:0060844 | smooth muscle tissue development(GO:0048745) arterial endothelial cell fate commitment(GO:0060844) |
| 0.8 | 2.3 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.8 | 3.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.7 | 5.1 | GO:0072132 | mesenchyme morphogenesis(GO:0072132) |
| 0.7 | 3.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.7 | 4.7 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.6 | 1.8 | GO:0048939 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.6 | 3.0 | GO:0048903 | anterior lateral line neuromast development(GO:0048901) anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.6 | 1.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.5 | 8.7 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.5 | 2.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.5 | 4.9 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.5 | 6.7 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.5 | 1.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.4 | 1.2 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.4 | 1.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.4 | 1.2 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.4 | 1.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.4 | 2.8 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.4 | 3.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.4 | 4.7 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.4 | 2.5 | GO:2000480 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 2.1 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.3 | 1.4 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.3 | 2.8 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.3 | 1.7 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.3 | 0.7 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.3 | 1.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 1.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.3 | 0.9 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.3 | 0.9 | GO:0030431 | sleep(GO:0030431) |
| 0.3 | 0.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.3 | 0.8 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.3 | 1.4 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 1.1 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.3 | 1.9 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.3 | 1.3 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.3 | 1.1 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.3 | 3.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.3 | 1.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 1.5 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.3 | 4.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 0.9 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 2.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 2.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 3.6 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.2 | 2.8 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.2 | 1.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 1.1 | GO:0090317 | negative regulation of intracellular protein transport(GO:0090317) |
| 0.2 | 2.0 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.2 | 0.6 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.2 | 1.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 0.5 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 1.6 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.2 | 2.9 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 0.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 2.7 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.2 | 0.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 0.8 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 3.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.7 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 2.5 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 4.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 1.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 2.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 2.3 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 1.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 2.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.6 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.1 | 5.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.6 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.7 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 3.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 2.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 2.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 2.9 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 3.3 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 3.1 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 0.5 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 1.3 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 3.8 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 0.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 1.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 4.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 0.4 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.1 | 1.6 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.1 | 3.8 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 2.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.3 | GO:0045428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.1 | 0.4 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 3.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.4 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 | 0.4 | GO:0021548 | pons development(GO:0021548) |
| 0.1 | 1.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 3.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.2 | GO:0048011 | neurotrophin signaling pathway(GO:0038179) neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 1.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.8 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 2.1 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 0.9 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.4 | GO:0060827 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 1.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.6 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 1.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 1.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 1.7 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 3.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.1 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.5 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) optokinetic behavior(GO:0007634) |
| 0.0 | 1.3 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 1.7 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.8 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.4 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.3 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 2.3 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 2.3 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.4 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 1.3 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.7 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.8 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.1 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.0 | 3.5 | GO:0042060 | wound healing(GO:0042060) |
| 0.0 | 0.7 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 1.6 | GO:0098609 | cell-cell adhesion(GO:0098609) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.7 | 4.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.5 | 2.7 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.4 | 5.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 2.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.4 | 2.1 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.4 | 1.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 2.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 2.9 | GO:1904949 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.3 | 4.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 1.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 2.8 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 1.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 2.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 2.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 3.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 3.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 3.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 1.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 3.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.4 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.1 | 2.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 7.5 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.1 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.7 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 4.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 1.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 3.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 3.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 2.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 3.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 7.2 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 4.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 3.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 20.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.0 | GO:0035770 | ribonucleoprotein granule(GO:0035770) cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 11.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 29.4 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.3 | GO:0005604 | basement membrane(GO:0005604) extracellular matrix component(GO:0044420) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.9 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 1.2 | GO:0045202 | synapse(GO:0045202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 1.2 | 3.6 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 1.1 | 4.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.9 | 3.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.8 | 1.6 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.6 | 3.4 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.6 | 1.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.5 | 2.1 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.5 | 4.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.5 | 2.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.5 | 1.9 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.4 | 1.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.4 | 1.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 2.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.4 | 4.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) fatty acid transporter activity(GO:0015245) |
| 0.3 | 1.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 2.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 1.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 1.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 1.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.3 | 2.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.8 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) C-C chemokine receptor activity(GO:0016493) |
| 0.3 | 4.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 3.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 2.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.2 | 2.3 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 2.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 1.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.2 | 1.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 0.9 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 0.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 2.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 0.9 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 8.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 2.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 0.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 2.0 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.2 | 1.1 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.2 | 1.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 0.5 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.2 | 4.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 1.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 4.7 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.2 | 3.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 4.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 2.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 2.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 2.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.3 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 3.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.2 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.1 | 5.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.5 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 2.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 5.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 1.7 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 2.3 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 1.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 5.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 2.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.7 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.6 | GO:0009931 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 2.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 31.8 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 6.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 1.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0000049 | tRNA binding(GO:0000049) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.6 | 4.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 3.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 7.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 4.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 3.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 1.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 3.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 5.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.1 | 5.6 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.3 | 2.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 3.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 2.3 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.2 | 1.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 1.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 1.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 4.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 5.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 4.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 2.3 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 1.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |