Project
DANIO-CODE
Navigation
Downloads
Results for zbtb16b
Z-value: 1.60
Transcription factors associated with zbtb16b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb16b
|
ENSDARG00000074526 | zinc finger and BTB domain containing 16b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb16b | dr10_dc_chr15_-_18425091_18425237 | -0.91 | 1.2e-06 | Click! |
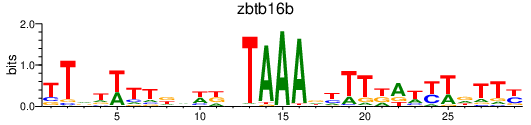
Activity profile of zbtb16b motif
Sorted Z-values of zbtb16b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb16b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_17695289 | 8.11 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr7_-_24567184 | 8.11 |
ENSDART00000131530
|
fam113
|
family with sequence similarity 113 |
| chr7_+_17695173 | 6.59 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr19_+_15536640 | 5.36 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr7_-_24604255 | 4.77 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr14_-_32535860 | 4.15 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr7_-_24249672 | 3.92 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr24_+_39285121 | 3.85 |
|
|
|
| chr19_-_28004606 | 3.82 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr19_-_4206333 | 3.51 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr14_-_6738260 | 3.41 |
ENSDART00000171792
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr21_-_43641419 | 3.31 |
ENSDART00000151486
ENSDART00000151115 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr16_+_47283253 | 3.28 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr21_-_21491268 | 3.21 |
ENSDART00000014254
|
diablob
|
diablo, IAP-binding mitochondrial protein b |
| chr17_+_16421892 | 3.07 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr14_+_24543399 | 2.96 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr21_+_3624059 | 2.91 |
ENSDART00000111699
|
tor1
|
torsin family 1 |
| chr4_-_2319514 | 2.78 |
ENSDART00000150039
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr7_-_24567002 | 2.75 |
ENSDART00000139455
|
fam113
|
family with sequence similarity 113 |
| chr20_-_22170411 | 2.72 |
ENSDART00000155568
|
BX088688.3
|
ENSDARG00000097598 |
| chr15_+_12504537 | 2.56 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr4_-_4603205 | 2.54 |
ENSDART00000130601
|
CABZ01020840.1
|
ENSDARG00000090401 |
| chr2_+_44659334 | 2.47 |
ENSDART00000155017
|
pask
|
PAS domain containing serine/threonine kinase |
| chr11_+_24583090 | 2.46 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr22_-_7748347 | 2.39 |
ENSDART00000097276
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr25_+_28419390 | 2.38 |
ENSDART00000154681
|
si:ch211-106e7.2
|
si:ch211-106e7.2 |
| chr23_-_27579257 | 2.36 |
ENSDART00000137229
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr16_+_47283374 | 2.36 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr21_-_9291200 | 2.24 |
ENSDART00000160932
|
sdad1
|
SDA1 domain containing 1 |
| chr5_+_4906690 | 2.19 |
ENSDART00000124022
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr15_-_25158558 | 2.13 |
ENSDART00000142609
|
exo5
|
exonuclease 5 |
| chr24_-_2454189 | 2.08 |
ENSDART00000093331
|
rreb1a
|
ras responsive element binding protein 1a |
| chr24_-_23570846 | 2.07 |
ENSDART00000084954
ENSDART00000004013 ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr7_-_24249630 | 2.06 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr1_+_43471754 | 2.05 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr16_+_41222408 | 2.05 |
ENSDART00000128963
|
nek11
|
NIMA-related kinase 11 |
| chr1_+_43471879 | 2.04 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr15_+_12504492 | 2.04 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr12_+_34157065 | 2.03 |
ENSDART00000153127
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr25_+_19443128 | 2.03 |
ENSDART00000141056
|
nedd1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr4_-_29095531 | 2.03 |
ENSDART00000142951
|
si:dkey-23a23.2
|
si:dkey-23a23.2 |
| chr21_-_2316052 | 2.00 |
ENSDART00000158459
|
zgc:193790
|
zgc:193790 |
| chr8_+_28715451 | 2.00 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr1_-_689420 | 1.98 |
|
|
|
| chr25_-_34599241 | 1.96 |
ENSDART00000156727
|
si:dkey-108k21.21
|
si:dkey-108k21.21 |
| chr19_-_4206143 | 1.96 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr2_-_7959150 | 1.94 |
ENSDART00000136074
|
b3gnt5b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5b |
| chr16_+_32775164 | 1.88 |
ENSDART00000125663
|
si:dkey-165n16.1
|
si:dkey-165n16.1 |
| chr5_-_45358634 | 1.87 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr14_-_23798158 | 1.87 |
ENSDART00000172747
|
si:ch211-277c7.7
|
si:ch211-277c7.7 |
| chr14_-_14353451 | 1.86 |
ENSDART00000170355
ENSDART00000159888 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr18_-_50439653 | 1.84 |
ENSDART00000151038
|
CU862078.1
|
ENSDARG00000096262 |
| chr1_+_43471798 | 1.81 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr5_+_6789490 | 1.81 |
ENSDART00000176668
ENSDART00000174471 |
CABZ01073148.1
|
ENSDARG00000106722 |
| chr3_+_62005461 | 1.81 |
ENSDART00000168710
|
znf1124
|
zinc finger protein 1124 |
| chr10_+_24721086 | 1.80 |
ENSDART00000079549
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr14_+_3399943 | 1.79 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr12_-_10327938 | 1.78 |
ENSDART00000007335
|
ndc80
|
NDC80 kinetochore complex component |
| chr21_+_18870471 | 1.77 |
ENSDART00000160185
|
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr18_+_8959686 | 1.76 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr4_-_65423861 | 1.74 |
ENSDART00000175471
|
BX511215.1
|
ENSDARG00000098062 |
| chr12_+_32316774 | 1.73 |
ENSDART00000153144
|
CT573006.1
|
ENSDARG00000096761 |
| chr9_-_8916789 | 1.72 |
ENSDART00000144673
|
rab20
|
RAB20, member RAS oncogene family |
| chr22_+_1153361 | 1.71 |
ENSDART00000159761
|
irf6
|
interferon regulatory factor 6 |
| chr4_-_4603294 | 1.70 |
ENSDART00000130601
|
CABZ01020840.1
|
ENSDARG00000090401 |
| chr22_-_7412566 | 1.69 |
ENSDART00000161615
ENSDART00000158210 |
BX511034.5
|
ENSDARG00000099295 |
| chr24_+_12691540 | 1.69 |
|
|
|
| chr9_-_40129231 | 1.68 |
ENSDART00000177531
|
CABZ01092969.1
|
ENSDARG00000108990 |
| chr8_-_31706973 | 1.67 |
ENSDART00000061832
|
si:dkey-46a10.3
|
si:dkey-46a10.3 |
| chr24_+_25995817 | 1.67 |
ENSDART00000139017
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr1_+_43471683 | 1.66 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr11_+_19440815 | 1.65 |
ENSDART00000005639
|
thoc7
|
THO complex 7 |
| chr21_+_599565 | 1.64 |
ENSDART00000177311
|
CABZ01072933.1
|
ENSDARG00000106379 |
| KN149859v1_+_8756 | 1.60 |
ENSDART00000157687
|
ENSDARG00000103399
|
ENSDARG00000103399 |
| KN150127v1_-_16267 | 1.59 |
|
|
|
| chr17_-_10581958 | 1.57 |
ENSDART00000051526
|
jmjd7
|
jumonji domain containing 7 |
| chr4_-_35856278 | 1.57 |
ENSDART00000162568
|
znf1125
|
zinc finger protein 1125 |
| chr3_-_26060098 | 1.57 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr15_-_17088606 | 1.56 |
ENSDART00000154719
ENSDART00000014465 |
hip1
|
huntingtin interacting protein 1 |
| chr10_+_34057791 | 1.56 |
ENSDART00000149934
|
kl
|
klotho |
| chr24_+_12691847 | 1.50 |
|
|
|
| chr24_+_36451347 | 1.50 |
ENSDART00000142264
|
grnb
|
granulin b |
| chr3_-_26060047 | 1.50 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr13_-_6150836 | 1.49 |
ENSDART00000092105
|
agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| KN149990v1_+_7315 | 1.49 |
|
|
|
| chr20_-_16271738 | 1.48 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr14_+_50232242 | 1.47 |
ENSDART00000110099
|
rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr7_+_39392939 | 1.39 |
ENSDART00000146171
|
zgc:158564
|
zgc:158564 |
| chr7_-_57055583 | 1.38 |
|
|
|
| chr10_-_21971084 | 1.38 |
ENSDART00000174954
|
CABZ01031892.1
|
ENSDARG00000107280 |
| chr22_-_20378023 | 1.38 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr25_-_27222696 | 1.38 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr2_-_30216703 | 1.37 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr4_-_36838449 | 1.35 |
|
|
|
| chr16_+_36118041 | 1.34 |
ENSDART00000166040
|
sh3bp5b
|
SH3-domain binding protein 5b (BTK-associated) |
| chr5_-_66614185 | 1.34 |
ENSDART00000147053
|
rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr15_-_25158683 | 1.33 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr16_+_32775303 | 1.32 |
ENSDART00000125663
|
si:dkey-165n16.1
|
si:dkey-165n16.1 |
| chr24_-_23973276 | 1.32 |
ENSDART00000111096
|
zgc:112982
|
zgc:112982 |
| chr11_-_22828072 | 1.32 |
ENSDART00000127791
|
atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr2_+_42923652 | 1.32 |
ENSDART00000168318
|
BX005083.1
|
ENSDARG00000097562 |
| chr6_-_60054220 | 1.28 |
ENSDART00000057466
|
ccndbp1
|
cyclin D-type binding-protein 1 |
| chr7_-_5172270 | 1.26 |
ENSDART00000172963
|
FP236789.1
|
ENSDARG00000105406 |
| chr21_+_19381959 | 1.25 |
ENSDART00000080110
|
amacr
|
alpha-methylacyl-CoA racemase |
| chr15_+_38319554 | 1.24 |
ENSDART00000122134
|
stim1a
|
stromal interaction molecule 1a |
| chr7_-_47990507 | 1.24 |
ENSDART00000135671
|
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr11_-_28508152 | 1.21 |
|
|
|
| chr24_+_25064535 | 1.21 |
ENSDART00000162134
ENSDART00000081043 |
zgc:174160
|
zgc:174160 |
| chr3_-_25017438 | 1.20 |
ENSDART00000089325
|
mief1
|
mitochondrial elongation factor 1 |
| chr21_-_16979447 | 1.19 |
|
|
|
| chr3_-_7709509 | 1.19 |
|
|
|
| chr16_+_14119910 | 1.19 |
ENSDART00000122272
|
fdps
|
farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr13_-_24298620 | 1.19 |
ENSDART00000125949
|
cep68
|
centrosomal protein 68 |
| chr16_+_54953101 | 1.19 |
|
|
|
| chr13_-_24298544 | 1.18 |
ENSDART00000147514
|
cep68
|
centrosomal protein 68 |
| chr25_-_5776878 | 1.18 |
ENSDART00000178323
|
CABZ01067754.1
|
ENSDARG00000107684 |
| chr14_-_8779307 | 1.16 |
ENSDART00000054710
|
polr1d
|
polymerase (RNA) I polypeptide D |
| chr3_-_52428451 | 1.14 |
ENSDART00000155248
|
si:dkey-210j14.5
|
si:dkey-210j14.5 |
| chr8_-_31706783 | 1.13 |
ENSDART00000061832
|
si:dkey-46a10.3
|
si:dkey-46a10.3 |
| chr22_-_6164703 | 1.13 |
ENSDART00000129829
|
si:rp71-36a1.5
|
si:rp71-36a1.5 |
| chr14_-_14353487 | 1.12 |
ENSDART00000172241
|
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr6_+_8916643 | 1.12 |
|
|
|
| chr24_-_23570895 | 1.11 |
ENSDART00000084954
ENSDART00000004013 ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr9_+_34423285 | 1.09 |
ENSDART00000174944
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr15_+_44120328 | 1.09 |
|
|
|
| chr1_+_43471906 | 1.09 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr13_-_694153 | 1.07 |
ENSDART00000102897
|
polh
|
polymerase (DNA directed), eta |
| chr25_+_18378608 | 1.05 |
ENSDART00000158142
ENSDART00000073563 |
tes
|
testis derived transcript (3 LIM domains) |
| chr1_-_37642238 | 1.05 |
ENSDART00000015323
|
fbxo8
|
F-box protein 8 |
| chr22_-_38472936 | 1.05 |
ENSDART00000098461
|
ptk7a
|
protein tyrosine kinase 7a |
| chr22_-_680961 | 1.05 |
ENSDART00000149320
|
arl8a
|
ADP-ribosylation factor-like 8A |
| chr23_+_25942056 | 1.04 |
ENSDART00000103934
|
ttpal
|
tocopherol (alpha) transfer protein-like |
| chr6_+_11445579 | 1.04 |
ENSDART00000151447
ENSDART00000151618 |
calcrlb
|
calcitonin receptor-like b |
| chr22_+_2483845 | 1.04 |
ENSDART00000170192
|
CU570894.1
|
ENSDARG00000103886 |
| chr6_-_33886299 | 1.03 |
ENSDART00000115409
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr11_+_23657658 | 1.03 |
ENSDART00000160394
|
CR354444.2
|
ENSDARG00000103549 |
| chr9_+_22015827 | 1.02 |
ENSDART00000101985
|
zgc:162944
|
zgc:162944 |
| KN150158v1_-_15806 | 1.02 |
ENSDART00000163880
|
CABZ01067681.1
|
ENSDARG00000103190 |
| chr22_-_5979881 | 1.01 |
|
|
|
| chr25_-_17282381 | 1.01 |
ENSDART00000178173
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr8_+_26064660 | 1.01 |
ENSDART00000099283
|
dalrd3
|
DALR anticodon binding domain containing 3 |
| chr23_-_16160099 | 1.00 |
ENSDART00000042469
ENSDART00000146605 |
mrgbp
|
MRG/MORF4L binding protein |
| chr2_-_15364526 | 0.99 |
ENSDART00000131835
|
hccsa.1
|
holocytochrome c synthase a |
| chr22_+_1153570 | 0.98 |
ENSDART00000157442
|
irf6
|
interferon regulatory factor 6 |
| chr13_+_39056287 | 0.98 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr21_-_9291452 | 0.98 |
ENSDART00000169275
|
sdad1
|
SDA1 domain containing 1 |
| chr17_-_12095843 | 0.97 |
ENSDART00000155545
|
ahctf1
|
AT hook containing transcription factor 1 |
| chr4_-_25443857 | 0.97 |
|
|
|
| chr11_-_7786415 | 0.95 |
ENSDART00000154569
|
CR450813.1
|
ENSDARG00000097035 |
| chr25_+_2968769 | 0.94 |
|
|
|
| chr11_-_411950 | 0.92 |
ENSDART00000137121
|
nuf2
|
NUF2, NDC80 kinetochore complex component, homolog |
| chr7_+_71585147 | 0.91 |
ENSDART00000161344
|
nsun6
|
NOL1/NOP2/Sun domain family, member 6 |
| chr20_+_2651918 | 0.88 |
ENSDART00000058777
|
slc35a1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr18_-_42072998 | 0.86 |
|
|
|
| chr21_-_2042057 | 0.86 |
ENSDART00000163608
|
add1
|
adducin 1 (alpha) |
| chr5_-_66614227 | 0.86 |
ENSDART00000147053
|
rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr8_-_6782167 | 0.86 |
ENSDART00000135834
ENSDART00000172157 |
dock5
|
dedicator of cytokinesis 5 |
| chr10_+_24721139 | 0.85 |
ENSDART00000144710
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr6_-_48062769 | 0.85 |
ENSDART00000145376
|
CR788236.1
|
ENSDARG00000094064 |
| chr12_-_13927800 | 0.85 |
ENSDART00000066368
|
klhl11
|
kelch-like family member 11 |
| KN149695v1_+_135563 | 0.85 |
|
|
|
| chr25_+_11212787 | 0.84 |
ENSDART00000159583
|
FQ312013.1
|
ENSDARG00000099473 |
| chr9_+_7829443 | 0.83 |
|
|
|
| chr8_+_28715581 | 0.83 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr10_-_7677571 | 0.79 |
ENSDART00000166234
|
ubxn8
|
UBX domain protein 8 |
| chr21_-_2322963 | 0.79 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr5_+_6391432 | 0.78 |
ENSDART00000170564
ENSDART00000086666 |
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr8_+_12913443 | 0.77 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr11_+_19208363 | 0.75 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr9_-_35073289 | 0.75 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr11_+_40847576 | 0.75 |
ENSDART00000041531
|
park7
|
parkinson protein 7 |
| chr18_+_15873032 | 0.74 |
ENSDART00000141800
|
eea1
|
early endosome antigen 1 |
| chr21_+_16979503 | 0.74 |
ENSDART00000065755
|
gpn3
|
GPN-loop GTPase 3 |
| chr23_-_38228511 | 0.73 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr16_+_13795633 | 0.72 |
ENSDART00000080342
|
josd2
|
Josephin domain containing 2 |
| chr22_-_4867420 | 0.72 |
ENSDART00000132942
|
ncl1
|
nicalin |
| chr22_+_9493668 | 0.71 |
ENSDART00000143953
|
strip1
|
striatin interacting protein 1 |
| chr21_-_13128373 | 0.71 |
ENSDART00000080347
ENSDART00000123238 |
wdr34
|
WD repeat domain 34 |
| chr4_-_43539382 | 0.71 |
|
|
|
| chr10_+_36495520 | 0.71 |
ENSDART00000124060
ENSDART00000043802 |
uspl1
|
ubiquitin specific peptidase like 1 |
| chr7_+_39393052 | 0.71 |
ENSDART00000146171
|
zgc:158564
|
zgc:158564 |
| chr16_+_14119938 | 0.71 |
ENSDART00000059928
|
fdps
|
farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr22_+_22412894 | 0.71 |
ENSDART00000089622
|
kif14
|
kinesin family member 14 |
| chr4_+_10002607 | 0.69 |
ENSDART00000148105
|
naf1
|
nuclear assembly factor 1 homolog (S. cerevisiae) |
| chr2_-_30216455 | 0.68 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr13_+_39056158 | 0.67 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr7_+_39393106 | 0.67 |
ENSDART00000047271
ENSDART00000113458 |
zgc:158564
|
zgc:158564 |
| chr12_-_34157019 | 0.66 |
ENSDART00000109196
|
pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr6_+_36494804 | 0.66 |
ENSDART00000138778
|
BX950188.1
|
ENSDARG00000094164 |
| chr2_-_55565099 | 0.66 |
ENSDART00000149062
|
rab8a
|
RAB8A, member RAS oncogene family |
| chr1_-_39801699 | 0.64 |
ENSDART00000114659
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr23_-_41929112 | 0.60 |
ENSDART00000131235
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr25_+_21618694 | 0.60 |
ENSDART00000148920
|
znf277
|
zinc finger protein 277 |
| chr12_+_3834709 | 0.59 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr12_+_22800197 | 0.59 |
ENSDART00000136042
|
rab18a
|
RAB18A, member RAS oncogene family |
| chr2_+_5929231 | 0.58 |
ENSDART00000136180
|
dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr16_-_40093241 | 0.57 |
ENSDART00000145278
|
si:dkey-29b11.3
|
si:dkey-29b11.3 |
| chr18_+_49974874 | 0.57 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr1_+_11707886 | 0.57 |
ENSDART00000008127
|
zgc:77739
|
zgc:77739 |
| chr23_-_5188577 | 0.56 |
ENSDART00000123191
|
ube2t
|
ubiquitin-conjugating enzyme E2T (putative) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.6 | 3.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.6 | 4.8 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.5 | 1.5 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.5 | 2.5 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.5 | 1.9 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.4 | 5.6 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.4 | 1.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.4 | 1.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.4 | 1.8 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.3 | 1.2 | GO:0051150 | regulation of smooth muscle cell differentiation(GO:0051150) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 | 2.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.3 | 8.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.3 | 1.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 1.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) |
| 0.2 | 0.7 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.2 | 3.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 1.0 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.2 | 0.7 | GO:0000493 | box H/ACA snoRNP assembly(GO:0000493) |
| 0.2 | 0.7 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 0.9 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) |
| 0.2 | 1.0 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.2 | 2.0 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 | 0.6 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.2 | 5.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 4.1 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.2 | 0.5 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 0.7 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 3.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 5.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.7 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.3 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 3.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.2 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 0.9 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 2.7 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 2.0 | GO:0097696 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.1 | 1.9 | GO:0007632 | visual behavior(GO:0007632) |
| 0.1 | 0.9 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 1.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 2.1 | GO:0051896 | regulation of protein kinase B signaling(GO:0051896) |
| 0.1 | 1.6 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 1.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:0021610 | facial nerve morphogenesis(GO:0021610) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 1.0 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.4 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 2.6 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 6.0 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.1 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.8 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 2.4 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.8 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 1.3 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 1.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.5 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 1.0 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.1 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.8 | GO:0001510 | RNA methylation(GO:0001510) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.5 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.7 | 2.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.7 | 2.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.6 | 1.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.6 | 2.8 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.3 | 1.6 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.3 | 2.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 1.6 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 2.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.0 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.6 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.8 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 9.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 6.0 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.9 | 3.5 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.8 | 4.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.7 | 3.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.5 | 1.5 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.4 | 3.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.3 | 2.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.3 | 1.3 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.3 | 2.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.3 | 2.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 0.8 | GO:0097642 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) calcitonin receptor activity(GO:0004948) calcitonin family receptor activity(GO:0097642) |
| 0.2 | 1.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 2.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 1.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 3.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 6.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.9 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.9 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 4.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.8 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 1.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 5.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 1.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0090482 | azole transporter activity(GO:0045118) vitamin transmembrane transporter activity(GO:0090482) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 3.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 9.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.0 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.4 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 2.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 2.0 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.3 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 2.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 4.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 3.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 4.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.6 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 4.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.0 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.8 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |