Project
DANIO-CODE
Navigation
Downloads
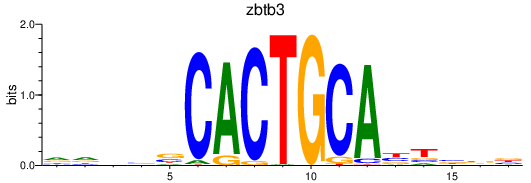
Results for zbtb3
Z-value: 0.74
Transcription factors associated with zbtb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb3
|
ENSDARG00000036235 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb3 | dr10_dc_chr14_+_35065189_35065257 | -0.55 | 2.6e-02 | Click! |
Activity profile of zbtb3 motif
Sorted Z-values of zbtb3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_3458086 | 2.16 |
ENSDART00000160977
ENSDART00000114168 |
CU469503.1
itga6a
|
ENSDARG00000099348 integrin, alpha 6a |
| chr16_-_42968807 | 1.79 |
ENSDART00000154757
|
txnipb
|
thioredoxin interacting protein b |
| chr3_-_60953138 | 1.49 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr7_-_35161302 | 1.49 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr14_+_6239755 | 1.43 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr9_+_53788118 | 1.41 |
ENSDART00000125715
ENSDART00000166582 ENSDART00000162224 |
dct
|
dopachrome tautomerase |
| chr16_-_42968860 | 1.33 |
ENSDART00000102345
|
txnipb
|
thioredoxin interacting protein b |
| chr9_-_32942783 | 1.29 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr7_-_25623974 | 1.29 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr9_-_34141609 | 1.22 |
|
|
|
| chr12_+_24833569 | 1.12 |
ENSDART00000014868
|
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr20_-_42821812 | 1.11 |
ENSDART00000138957
|
gpr31
|
G protein-coupled receptor 31 |
| chr7_-_25816549 | 1.09 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr14_+_24637864 | 1.04 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr7_-_25816655 | 0.98 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr16_+_10531986 | 0.97 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr22_-_36552513 | 0.95 |
ENSDART00000129318
|
CABZ01045212.1
|
ENSDARG00000087525 |
| chr17_+_34168095 | 0.93 |
|
|
|
| chr23_+_6298911 | 0.92 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr22_+_528076 | 0.91 |
|
|
|
| chr20_+_25441689 | 0.89 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr22_-_29387056 | 0.84 |
ENSDART00000121599
|
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr8_-_12171880 | 0.84 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr1_+_50277442 | 0.82 |
ENSDART00000150420
|
otx1a
|
orthodenticle homeobox 1a |
| chr3_+_24327586 | 0.81 |
ENSDART00000153551
|
cbx6b
|
chromobox homolog 6b |
| chr11_-_23151247 | 0.81 |
|
|
|
| chr17_-_45577704 | 0.78 |
ENSDART00000109831
|
BX088653.1
|
ENSDARG00000074434 |
| chr16_+_10531926 | 0.75 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr16_-_32059848 | 0.73 |
ENSDART00000138701
|
gstk1
|
glutathione S-transferase kappa 1 |
| chr1_-_53746151 | 0.72 |
ENSDART00000179565
|
CABZ01043826.1
|
ENSDARG00000108130 |
| chr12_-_36191048 | 0.72 |
ENSDART00000177986
|
cep131
|
centrosomal protein 131 |
| chr8_-_12171755 | 0.72 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr5_+_19737802 | 0.70 |
ENSDART00000153643
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr10_+_32007448 | 0.68 |
ENSDART00000019416
|
lhfp
|
lipoma HMGIC fusion partner |
| chr1_+_44113013 | 0.66 |
ENSDART00000059227
|
ndufs8a
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8a |
| chr2_+_47885551 | 0.66 |
|
|
|
| chr13_-_2083873 | 0.63 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr18_+_16948165 | 0.61 |
|
|
|
| chr9_-_28464236 | 0.59 |
ENSDART00000146932
|
creb1b
|
cAMP responsive element binding protein 1b |
| chr13_+_23152038 | 0.58 |
ENSDART00000171676
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr8_+_26103807 | 0.58 |
|
|
|
| chr21_+_45325383 | 0.55 |
ENSDART00000029946
|
ube2b
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog) |
| chr7_-_25816498 | 0.53 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr17_-_36894361 | 0.52 |
ENSDART00000176989
|
myo6b
|
myosin VIb |
| chr18_+_37034218 | 0.51 |
|
|
|
| chr1_+_44141337 | 0.51 |
|
|
|
| chr6_+_58704352 | 0.50 |
ENSDART00000172466
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr20_+_38768950 | 0.48 |
ENSDART00000146544
|
mpv17
|
MpV17 mitochondrial inner membrane protein |
| chr18_+_48434068 | 0.48 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| KN150171v1_+_3909 | 0.48 |
ENSDART00000171048
|
CABZ01071639.1
|
ENSDARG00000099435 |
| chr1_-_44002772 | 0.48 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr25_+_13566205 | 0.47 |
ENSDART00000008989
|
ccdc113
|
coiled-coil domain containing 113 |
| KN149781v1_+_4885 | 0.47 |
ENSDART00000165339
|
CDC37
|
cell division cycle 37 |
| chr19_+_31944725 | 0.45 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr21_-_45796826 | 0.44 |
|
|
|
| chr19_-_2084281 | 0.43 |
|
|
|
| chr16_-_24746483 | 0.43 |
ENSDART00000058959
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr6_-_39009073 | 0.41 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr25_-_35037680 | 0.38 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr16_-_24682800 | 0.37 |
ENSDART00000156407
ENSDART00000154072 |
si:dkey-56f14.4
|
si:dkey-56f14.4 |
| chr14_-_1342450 | 0.34 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr22_-_11106940 | 0.33 |
ENSDART00000016873
|
atp6ap2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr16_-_12095302 | 0.32 |
ENSDART00000148666
ENSDART00000029121 |
usp5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr14_-_33518100 | 0.32 |
ENSDART00000163877
|
ocrl
|
oculocerebrorenal syndrome of Lowe |
| chr24_+_6078848 | 0.31 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr14_-_23503974 | 0.30 |
ENSDART00000054264
|
nr3c1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr8_+_30912152 | 0.29 |
ENSDART00000171400
|
derl3
|
derlin 3 |
| chr23_-_26961559 | 0.29 |
ENSDART00000038491
|
adcy6b
|
adenylate cyclase 6b |
| chr13_+_30968221 | 0.26 |
ENSDART00000014922
|
arhgap22
|
Rho GTPase activating protein 22 |
| chr16_+_4865780 | 0.23 |
ENSDART00000039054
|
mecr
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr8_-_8407754 | 0.21 |
ENSDART00000137382
|
cdk16
|
cyclin-dependent kinase 16 |
| chr9_+_14052160 | 0.21 |
ENSDART00000124267
|
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr14_-_2873620 | 0.21 |
|
|
|
| chr8_+_17493410 | 0.20 |
ENSDART00000147760
|
si:ch73-70k4.1
|
si:ch73-70k4.1 |
| chr16_-_27289365 | 0.20 |
ENSDART00000126347
|
alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr13_+_23152160 | 0.20 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr8_-_8408046 | 0.18 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr24_+_6078791 | 0.18 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr3_+_52985863 | 0.17 |
ENSDART00000031234
|
stxbp2
|
syntaxin binding protein 2 |
| chr17_+_52438099 | 0.17 |
ENSDART00000135246
|
dlst
|
dihydrolipoamide S-succinyltransferase |
| chr11_+_38858351 | 0.17 |
ENSDART00000155746
|
cdc42
|
cell division cycle 42 |
| chr2_+_58658858 | 0.16 |
|
|
|
| chr6_+_12805091 | 0.13 |
ENSDART00000091700
|
catip
|
ciliogenesis associated TTC17 interacting protein |
| chr14_+_1342695 | 0.11 |
|
|
|
| chr7_-_25624212 | 0.11 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr16_-_24746425 | 0.09 |
ENSDART00000058959
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr7_-_25624128 | 0.06 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr6_-_39008833 | 0.06 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr14_-_33517822 | 0.05 |
ENSDART00000112268
|
ocrl
|
oculocerebrorenal syndrome of Lowe |
| chr5_+_61136262 | 0.05 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr8_-_52729505 | 0.04 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr23_+_24711233 | 0.04 |
|
|
|
| chr11_+_31362089 | 0.03 |
ENSDART00000158494
ENSDART00000173083 |
FO704610.1
|
ENSDARG00000100400 |
| chr9_-_30692456 | 0.02 |
|
|
|
| chr9_-_28464146 | 0.01 |
ENSDART00000137582
|
creb1b
|
cAMP responsive element binding protein 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0006581 | response to amphetamine(GO:0001975) acetylcholine catabolic process(GO:0006581) response to amine(GO:0014075) |
| 0.4 | 1.8 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.4 | 1.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.3 | 1.6 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 0.5 | GO:0046048 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.2 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.5 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 0.7 | GO:0090317 | negative regulation of intracellular protein transport(GO:0090317) |
| 0.1 | 0.8 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.4 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 1.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) cellular response to cortisol stimulus(GO:0071387) |
| 0.1 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.7 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.5 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 2.3 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 1.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.0 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.5 | GO:0060119 | inner ear receptor cell development(GO:0060119) |
| 0.0 | 1.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.5 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.7 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 0.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.7 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.5 | GO:0030017 | sarcomere(GO:0030017) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0004104 | acetylcholinesterase activity(GO:0003990) cholinesterase activity(GO:0004104) |
| 0.3 | 1.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 1.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 0.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 3.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |