Project
DANIO-CODE
Navigation
Downloads
Results for zbtb38
Z-value: 0.71
Transcription factors associated with zbtb38
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb38
|
ENSDARG00000100607 | zinc finger and BTB domain containing 38 |
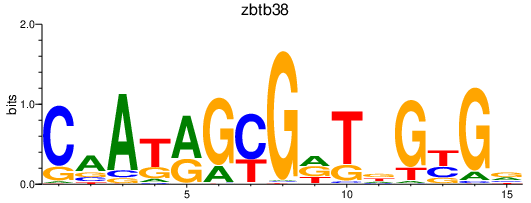
Activity profile of zbtb38 motif
Sorted Z-values of zbtb38 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb38
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_17163526 | 2.71 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr7_-_6273677 | 2.05 |
ENSDART00000173419
|
si:ch73-368j24.1
|
si:ch73-368j24.1 |
| chr14_-_27040866 | 2.00 |
ENSDART00000173053
|
CU062454.1
|
ENSDARG00000105327 |
| chr11_-_27715592 | 1.89 |
ENSDART00000168338
|
ece1
|
endothelin converting enzyme 1 |
| chr10_-_17214368 | 1.37 |
|
|
|
| chr16_-_23885481 | 1.36 |
ENSDART00000139964
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr14_-_47210912 | 1.28 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr3_-_9755642 | 1.21 |
ENSDART00000168366
|
crebbpb
|
CREB binding protein b |
| chr1_-_43347290 | 1.13 |
ENSDART00000073746
|
si:ch73-109d9.2
|
si:ch73-109d9.2 |
| chr22_+_26270259 | 1.08 |
ENSDART00000060898
|
mrps28
|
mitochondrial ribosomal protein S28 |
| chr22_+_26840517 | 1.07 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr22_-_16416848 | 1.02 |
ENSDART00000006290
|
plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr24_-_32607801 | 1.02 |
ENSDART00000143781
|
EIF1B
|
eukaryotic translation initiation factor 1B |
| chr16_+_43464824 | 1.01 |
ENSDART00000032778
|
rnf144b
|
ring finger protein 144B |
| chr22_+_1894340 | 1.01 |
ENSDART00000164158
|
znf1156
|
zinc finger protein 1156 |
| chr11_+_44922796 | 0.95 |
ENSDART00000172238
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr3_+_1451017 | 0.93 |
ENSDART00000161945
|
rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr13_-_44885278 | 0.90 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr23_+_36209868 | 0.87 |
ENSDART00000131711
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr16_+_29651573 | 0.83 |
ENSDART00000149520
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr9_+_42656327 | 0.82 |
|
|
|
| chr24_+_12693997 | 0.80 |
|
|
|
| chr15_-_8541318 | 0.79 |
ENSDART00000140213
|
npas1
|
neuronal PAS domain protein 1 |
| chr6_-_2037693 | 0.72 |
ENSDART00000159957
|
PXMP4
|
peroxisomal membrane protein 4 |
| chr3_+_40654510 | 0.71 |
ENSDART00000014729
|
arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr23_+_36209604 | 0.71 |
ENSDART00000134607
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr4_-_75558337 | 0.70 |
ENSDART00000174184
|
zgc:173770
|
zgc:173770 |
| chr10_+_1653720 | 0.70 |
ENSDART00000018532
|
triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr22_-_10509348 | 0.67 |
ENSDART00000131217
|
ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr16_+_23884575 | 0.67 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr1_-_43347586 | 0.66 |
ENSDART00000073746
|
si:ch73-109d9.2
|
si:ch73-109d9.2 |
| chr23_+_36209223 | 0.65 |
ENSDART00000134607
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr3_-_43134137 | 0.63 |
ENSDART00000165592
|
rmi2
|
RecQ mediated genome instability 2 |
| chr22_+_10202801 | 0.61 |
ENSDART00000105900
|
si:dkeyp-87e7.4
|
si:dkeyp-87e7.4 |
| chr8_-_4517590 | 0.60 |
ENSDART00000090731
|
dhx37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chr5_+_29250823 | 0.60 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr13_-_44885345 | 0.56 |
ENSDART00000074787
ENSDART00000125633 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr23_+_36209696 | 0.55 |
ENSDART00000134607
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr8_-_11164322 | 0.50 |
ENSDART00000147817
|
fam208b
|
family with sequence similarity 208, member B |
| chr13_-_44883668 | 0.49 |
|
|
|
| chr22_-_9796735 | 0.48 |
ENSDART00000144395
|
BX539307.3
|
ENSDARG00000093730 |
| chr13_+_18377514 | 0.48 |
|
|
|
| chr13_-_2812369 | 0.45 |
|
|
|
| chr13_-_8979137 | 0.44 |
ENSDART00000058056
|
mrps26
|
mitochondrial ribosomal protein S26 |
| chr16_-_22107826 | 0.43 |
ENSDART00000165824
ENSDART00000170634 |
setdb1b
|
SET domain, bifurcated 1b |
| chr12_-_49026424 | 0.41 |
|
|
|
| chr14_-_27041347 | 0.40 |
ENSDART00000173053
|
CU062454.1
|
ENSDARG00000105327 |
| chr10_+_1653868 | 0.40 |
ENSDART00000018532
|
triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr22_+_34726990 | 0.39 |
ENSDART00000082066
|
atpv0e2
|
ATPase, H+ transporting V0 subunit e2 |
| chr13_-_35851162 | 0.38 |
ENSDART00000075299
|
zgc:153911
|
zgc:153911 |
| chr16_+_43464694 | 0.37 |
ENSDART00000032778
|
rnf144b
|
ring finger protein 144B |
| chr13_-_35634988 | 0.36 |
ENSDART00000127476
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr5_+_53229583 | 0.36 |
ENSDART00000171107
|
naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr20_-_23327219 | 0.35 |
ENSDART00000142721
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr8_-_44941251 | 0.35 |
|
|
|
| chr15_-_17163487 | 0.34 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr21_+_1719308 | 0.31 |
|
|
|
| chr18_+_22285328 | 0.29 |
|
|
|
| KN150703v1_-_1874 | 0.29 |
|
|
|
| chr15_-_46792766 | 0.29 |
|
|
|
| chr1_-_24453469 | 0.27 |
ENSDART00000132355
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr13_-_44883880 | 0.25 |
|
|
|
| chr22_-_10509388 | 0.24 |
ENSDART00000131217
|
ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr3_+_40654220 | 0.23 |
ENSDART00000014729
|
arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr22_-_3165441 | 0.22 |
ENSDART00000158009
|
lonp1
|
lon peptidase 1, mitochondrial |
| chr5_-_51363461 | 0.20 |
ENSDART00000051003
|
cdk7
|
cyclin-dependent kinase 7 |
| chr8_-_31374705 | 0.20 |
ENSDART00000162872
|
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr16_+_29651305 | 0.18 |
ENSDART00000124232
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr5_+_71409912 | 0.17 |
ENSDART00000165835
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr11_+_11050173 | 0.17 |
|
|
|
| chr24_-_25316192 | 0.16 |
ENSDART00000105820
|
mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr13_+_29161591 | 0.16 |
ENSDART00000115023
|
PARG
|
poly(ADP-ribose) glycohydrolase |
| chr20_+_12809388 | 0.16 |
ENSDART00000163499
|
zgc:153383
|
zgc:153383 |
| chr14_-_47211080 | 0.14 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr7_-_27527490 | 0.12 |
ENSDART00000089640
|
PDE3B
|
phosphodiesterase 3B |
| chr11_+_13151425 | 0.12 |
|
|
|
| chr16_-_22107906 | 0.10 |
ENSDART00000165824
ENSDART00000170634 |
setdb1b
|
SET domain, bifurcated 1b |
| chr22_-_9796789 | 0.09 |
ENSDART00000144395
|
BX539307.3
|
ENSDARG00000093730 |
| chr2_-_57014322 | 0.08 |
ENSDART00000159480
|
btbd2b
|
BTB (POZ) domain containing 2b |
| chr20_+_28901045 | 0.07 |
ENSDART00000153046
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr5_-_13145749 | 0.07 |
ENSDART00000166957
|
purba
|
purine-rich element binding protein Ba |
| chr7_+_73634380 | 0.06 |
ENSDART00000126439
|
hist1h4l
|
histone 1, H4, like |
| chr18_-_5004815 | 0.04 |
|
|
|
| chr16_-_23885441 | 0.04 |
ENSDART00000139964
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr8_-_11164125 | 0.04 |
ENSDART00000147817
|
fam208b
|
family with sequence similarity 208, member B |
| chr2_+_1825714 | 0.02 |
ENSDART00000148624
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr10_+_18921391 | 0.01 |
ENSDART00000024127
|
ppp2r2ab
|
protein phosphatase 2, regulatory subunit B, alpha b |
| chr9_+_32367808 | 0.01 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.2 | 1.0 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 0.7 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 1.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 0.9 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.1 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 2.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.0 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.5 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.4 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.4 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.3 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.4 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 2.3 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 1.8 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 2.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 3.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |