Project
DANIO-CODE
Navigation
Downloads
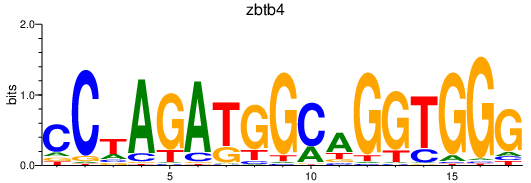
Results for zbtb4
Z-value: 0.24
Transcription factors associated with zbtb4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb4
|
ENSDARG00000105255 | zinc finger and BTB domain containing 4 |
Activity profile of zbtb4 motif
Sorted Z-values of zbtb4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_24770873 | 0.45 |
ENSDART00000165314
|
AL954715.1
|
ENSDARG00000103682 |
| chr16_+_17069742 | 0.30 |
ENSDART00000132150
|
rpl18
|
ribosomal protein L18 |
| chr14_+_6639864 | 0.20 |
ENSDART00000061001
|
gnb2l1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr2_+_7203572 | 0.17 |
ENSDART00000022963
|
cdc14aa
|
cell division cycle 14Aa |
| chr7_-_72127164 | 0.17 |
ENSDART00000176069
|
CABZ01057364.1
|
ENSDARG00000106736 |
| chr13_-_31516518 | 0.15 |
ENSDART00000172375
ENSDART00000125987 ENSDART00000143903 ENSDART00000158719 |
six4a
|
SIX homeobox 4a |
| chr8_+_25060320 | 0.14 |
ENSDART00000000744
|
sypl2b
|
synaptophysin-like 2b |
| chr23_+_31318844 | 0.11 |
ENSDART00000146510
|
irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr2_+_27000558 | 0.11 |
ENSDART00000099208
|
asph
|
aspartate beta-hydroxylase |
| chr4_+_5733160 | 0.08 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr7_+_6620497 | 0.07 |
ENSDART00000173130
|
sf3b2
|
splicing factor 3b, subunit 2 |
| chr16_-_32718222 | 0.07 |
ENSDART00000161395
|
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr7_-_17527827 | 0.07 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| KN150366v1_-_574 | 0.06 |
|
|
|
| chr13_-_31516399 | 0.06 |
ENSDART00000172375
ENSDART00000125987 ENSDART00000143903 ENSDART00000158719 |
six4a
|
SIX homeobox 4a |
| chr14_+_28126739 | 0.04 |
ENSDART00000026846
|
pin4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr14_+_28126352 | 0.04 |
ENSDART00000026846
|
pin4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr22_-_9790451 | 0.04 |
|
|
|
| chr15_-_28267088 | 0.03 |
ENSDART00000004200
|
sarm1
|
sterile alpha and TIR motif containing 1 |
| chr1_+_40763274 | 0.03 |
ENSDART00000135280
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr1_+_40762892 | 0.03 |
ENSDART00000010211
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr14_-_28126346 | 0.03 |
ENSDART00000054062
|
nek12
|
NIMA-related kinase 12 |
| chr8_+_25060389 | 0.03 |
ENSDART00000000744
|
sypl2b
|
synaptophysin-like 2b |
| chr8_+_39400230 | 0.03 |
|
|
|
| chr10_+_44847446 | 0.01 |
|
|
|
| chr23_+_35660991 | 0.00 |
ENSDART00000047082
|
gdap1l1
|
ganglioside induced differentiation associated protein 1-like 1 |
| chr10_+_44846866 | 0.00 |
|
|
|
| chr21_-_32244805 | 0.00 |
ENSDART00000178765
|
BX927388.2
|
ENSDARG00000107266 |
| chr1_+_1964902 | 0.00 |
ENSDART00000138396
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr4_-_66538843 | 0.00 |
ENSDART00000165173
|
BX548011.1
|
ENSDARG00000103357 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.0 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |