Project
DANIO-CODE
Navigation
Downloads
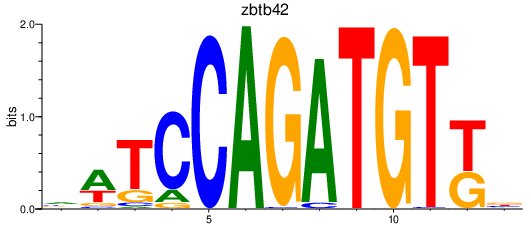
Results for zbtb42
Z-value: 1.78
Transcription factors associated with zbtb42
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb42
|
ENSDARG00000102761 | zinc finger and BTB domain containing 42 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb42 | dr10_dc_chr17_-_756401_756487 | 0.93 | 2.0e-07 | Click! |
Activity profile of zbtb42 motif
Sorted Z-values of zbtb42 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb42
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_5781358 | 6.05 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr16_-_26803204 | 5.94 |
ENSDART00000162665
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr9_-_33584190 | 5.66 |
ENSDART00000138844
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr15_-_14616083 | 4.78 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr7_+_34018862 | 4.72 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr13_+_11912981 | 4.48 |
ENSDART00000158244
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr15_+_34131126 | 4.46 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr1_-_11286438 | 4.42 |
ENSDART00000159981
ENSDART00000066638 |
grk4
|
G protein-coupled receptor kinase 4 |
| chr20_+_2443682 | 4.38 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr13_-_13163676 | 4.36 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr5_-_13872815 | 4.08 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr16_+_42925950 | 4.05 |
ENSDART00000159730
ENSDART00000014956 |
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr12_-_34614931 | 4.02 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr9_+_7609372 | 3.99 |
|
|
|
| chr10_-_11806373 | 3.96 |
ENSDART00000009715
|
adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr24_-_1307905 | 3.89 |
ENSDART00000159212
|
nrp1a
|
neuropilin 1a |
| chr4_-_2615160 | 3.84 |
ENSDART00000140760
|
e2f7
|
E2F transcription factor 7 |
| chr15_-_4322875 | 3.71 |
ENSDART00000173311
|
si:ch211-117a13.2
|
si:ch211-117a13.2 |
| chr9_-_9864049 | 3.69 |
ENSDART00000121456
|
fstl1b
|
follistatin-like 1b |
| chr7_-_25623974 | 3.69 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr7_+_23306546 | 3.58 |
ENSDART00000049885
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
| chr22_-_11712371 | 3.49 |
|
|
|
| chr23_+_21546553 | 3.41 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr4_-_76594677 | 3.40 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr10_-_22834248 | 3.39 |
ENSDART00000079469
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr21_-_20305406 | 3.29 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr17_+_29328653 | 3.24 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr2_+_11902170 | 3.15 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr17_-_14551031 | 3.02 |
ENSDART00000162452
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr9_+_7609527 | 3.01 |
|
|
|
| chr1_+_31131899 | 2.97 |
ENSDART00000075260
|
inab
|
internexin neuronal intermediate filament protein, alpha b |
| chr19_+_47719438 | 2.94 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr3_-_39245184 | 2.91 |
|
|
|
| chr22_+_17091411 | 2.82 |
ENSDART00000170076
|
frem1b
|
Fras1 related extracellular matrix 1b |
| chr2_+_31399487 | 2.71 |
ENSDART00000086863
|
colec12
|
collectin sub-family member 12 |
| chr23_+_36010592 | 2.69 |
ENSDART00000137507
|
hoxc3a
|
homeo box C3a |
| chr13_-_13163801 | 2.69 |
ENSDART00000156968
|
fgfr3
|
fibroblast growth factor receptor 3 |
| chr19_-_22894525 | 2.61 |
ENSDART00000090679
|
pleca
|
plectin a |
| KN150108v1_+_18276 | 2.59 |
|
|
|
| chr11_-_42826276 | 2.54 |
|
|
|
| chr13_-_25067585 | 2.52 |
ENSDART00000159585
|
adka
|
adenosine kinase a |
| chr10_+_7043530 | 2.52 |
|
|
|
| chr4_+_4223606 | 2.43 |
|
|
|
| chr19_+_5399813 | 2.42 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr16_-_13704905 | 2.36 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr18_-_46243753 | 2.31 |
ENSDART00000145999
|
prx
|
periaxin |
| chr7_+_5372830 | 2.28 |
ENSDART00000135271
|
MYADM
|
myeloid associated differentiation marker |
| chr25_-_4376129 | 2.25 |
|
|
|
| chr11_-_42826308 | 2.25 |
|
|
|
| chr12_+_22734974 | 2.19 |
ENSDART00000130594
|
afap1
|
actin filament associated protein 1 |
| chr21_-_18969970 | 2.14 |
ENSDART00000080269
|
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr14_-_35332747 | 2.11 |
ENSDART00000074710
|
pdgfc
|
platelet derived growth factor c |
| chr24_+_28364360 | 2.10 |
ENSDART00000018037
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr7_+_28862183 | 2.06 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr17_+_11353143 | 2.06 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr5_-_19357961 | 2.05 |
ENSDART00000088904
ENSDART00000163701 |
gltpa
|
glycolipid transfer protein a |
| chr5_-_33169062 | 2.04 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| chr12_+_13614102 | 2.02 |
ENSDART00000152689
|
oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr1_+_58029993 | 2.00 |
|
|
|
| chr3_+_17387551 | 1.97 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr20_+_14979154 | 1.96 |
ENSDART00000118157
|
dre-mir-199-1
|
dre-mir-199-1 |
| chr3_-_32464649 | 1.96 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr17_-_756401 | 1.94 |
ENSDART00000157622
|
zbtb42
|
zinc finger and BTB domain containing 42 |
| chr25_+_34997593 | 1.93 |
ENSDART00000058443
|
fibina
|
fin bud initiation factor a |
| chr4_-_76594399 | 1.91 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr3_-_54414513 | 1.91 |
ENSDART00000053106
|
s1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr16_-_20901879 | 1.84 |
ENSDART00000103630
|
creb5b
|
cAMP responsive element binding protein 5b |
| chr20_+_23339698 | 1.81 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr12_+_27612667 | 1.77 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr20_-_40553998 | 1.76 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr12_+_20578266 | 1.75 |
ENSDART00000016099
|
ENSDARG00000043818
|
ENSDARG00000043818 |
| chr2_+_5881998 | 1.75 |
ENSDART00000158045
ENSDART00000028703 |
slc35d1b
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1b |
| chr17_-_35004687 | 1.71 |
|
|
|
| chr1_+_44660098 | 1.67 |
ENSDART00000145486
|
si:ch211-243a20.3
|
si:ch211-243a20.3 |
| chr9_+_3416651 | 1.63 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr17_+_45961986 | 1.60 |
|
|
|
| chr10_+_20112765 | 1.57 |
ENSDART00000027612
|
xpo7
|
exportin 7 |
| chr20_-_47576935 | 1.55 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr13_+_18414806 | 1.54 |
ENSDART00000110197
|
zgc:154058
|
zgc:154058 |
| chr4_+_22754618 | 1.54 |
|
|
|
| chr9_-_31079647 | 1.53 |
|
|
|
| chr10_-_30016761 | 1.48 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr3_+_19582280 | 1.46 |
ENSDART00000109409
|
mrc2
|
mannose receptor, C type 2 |
| chr2_-_37061470 | 1.46 |
ENSDART00000056514
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr19_+_5399877 | 1.46 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr19_-_44495755 | 1.44 |
ENSDART00000132800
|
rad21b
|
RAD21 cohesin complex component b |
| chr10_-_24401876 | 1.42 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr20_+_6600246 | 1.42 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr16_+_9718837 | 1.41 |
ENSDART00000047920
|
enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr10_+_9134634 | 1.38 |
ENSDART00000110443
|
fstb
|
follistatin b |
| chr18_+_512813 | 1.37 |
ENSDART00000162513
|
gpib
|
glucose-6-phosphate isomerase b |
| chr23_-_27930478 | 1.37 |
ENSDART00000043462
|
acvrl1
|
activin A receptor type II-like 1 |
| chr17_+_38307512 | 1.36 |
ENSDART00000005296
|
nkx2.9
|
NK2 transcription factor related, locus 9 (Drosophila) |
| chr9_+_22953351 | 1.35 |
ENSDART00000090875
|
tnfaip6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr20_+_6600184 | 1.35 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr23_+_41295092 | 1.34 |
ENSDART00000109567
|
nhsa
|
Nance-Horan syndrome a (congenital cataracts and dental anomalies) |
| chr20_+_51034244 | 1.34 |
|
|
|
| chr14_-_45887648 | 1.34 |
ENSDART00000074208
|
cltb
|
clathrin, light chain B |
| chr12_-_28766713 | 1.33 |
ENSDART00000148459
|
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr19_-_20508993 | 1.28 |
ENSDART00000129917
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr8_+_39762186 | 1.28 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr18_+_48959401 | 1.27 |
ENSDART00000158104
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr1_+_58030069 | 1.27 |
|
|
|
| chr19_+_19916407 | 1.25 |
ENSDART00000166755
|
tril
|
TLR4 interactor with leucine-rich repeats |
| chr17_-_20267580 | 1.24 |
ENSDART00000078703
|
add3b
|
adducin 3 (gamma) b |
| chr3_-_36697213 | 1.24 |
ENSDART00000154553
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr20_+_29685096 | 1.23 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr6_+_39838929 | 1.19 |
ENSDART00000112637
|
smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr23_+_41129663 | 1.17 |
|
|
|
| chr24_+_34227202 | 1.16 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr2_-_24947660 | 1.13 |
ENSDART00000113356
ENSDART00000163038 |
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr23_+_20050920 | 1.13 |
ENSDART00000144790
|
cfap126
|
cilia and flagella associated protein 126 |
| chr24_-_28166614 | 1.11 |
ENSDART00000167861
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr14_-_23501428 | 1.06 |
|
|
|
| chr22_-_29133832 | 1.04 |
ENSDART00000172576
|
cbx6a
|
chromobox homolog 6a |
| chr19_+_47719402 | 1.03 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr24_+_30343687 | 1.02 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr6_-_43273456 | 1.01 |
|
|
|
| chr10_-_20524670 | 1.00 |
|
|
|
| chr24_-_25021505 | 0.99 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr2_-_44893769 | 0.99 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr18_+_48959017 | 0.98 |
ENSDART00000158104
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr19_-_36009321 | 0.98 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr16_-_26803131 | 0.96 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr9_+_32547244 | 0.95 |
ENSDART00000144608
|
plcl1
|
phospholipase C-like 1 |
| chr5_-_13872772 | 0.94 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr24_-_1308047 | 0.93 |
ENSDART00000164904
|
nrp1a
|
neuropilin 1a |
| chr10_-_20524586 | 0.90 |
|
|
|
| chr13_-_13163647 | 0.89 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr14_+_15698814 | 0.84 |
|
|
|
| chr4_+_12613575 | 0.72 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr5_-_21578927 | 0.68 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr16_-_51087813 | 0.68 |
ENSDART00000165408
|
si:dkeyp-97a10.3
|
si:dkeyp-97a10.3 |
| chr24_-_37989964 | 0.65 |
ENSDART00000128574
|
tmem204
|
transmembrane protein 204 |
| chr5_-_18510716 | 0.64 |
ENSDART00000002624
ENSDART00000110553 |
ranbp1
|
RAN binding protein 1 |
| chr5_-_38873781 | 0.64 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr11_-_27970634 | 0.63 |
|
|
|
| chr7_-_38340949 | 0.63 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr25_-_31049247 | 0.58 |
ENSDART00000009126
|
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr22_+_17581859 | 0.54 |
ENSDART00000035670
|
polr2eb
|
polymerase (RNA) II (DNA directed) polypeptide E, b |
| chr23_-_27930393 | 0.51 |
ENSDART00000043462
|
acvrl1
|
activin A receptor type II-like 1 |
| chr18_+_45553846 | 0.50 |
ENSDART00000138075
|
kifc3
|
kinesin family member C3 |
| chr5_-_41044841 | 0.50 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr1_-_17894339 | 0.48 |
ENSDART00000131548
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr20_+_14978952 | 0.46 |
ENSDART00000118157
|
dre-mir-199-1
|
dre-mir-199-1 |
| chr6_+_28232386 | 0.45 |
ENSDART00000163057
|
FP101882.1
|
ENSDARG00000098121 |
| chr22_+_25570287 | 0.44 |
|
|
|
| chr4_-_9608687 | 0.43 |
ENSDART00000067188
|
dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr13_+_11304073 | 0.43 |
ENSDART00000169895
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr2_-_52041699 | 0.41 |
ENSDART00000165568
|
pigr
|
polymeric immunoglobulin receptor |
| chr5_-_51352096 | 0.41 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr25_-_11287103 | 0.40 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr10_-_2944221 | 0.38 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr10_+_28700844 | 0.38 |
ENSDART00000047776
|
cblb
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr6_-_45993454 | 0.38 |
ENSDART00000154392
|
ca16b
|
carbonic anhydrase XVI b |
| chr13_+_30968221 | 0.36 |
ENSDART00000014922
|
arhgap22
|
Rho GTPase activating protein 22 |
| chr2_-_37061049 | 0.35 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr23_-_439495 | 0.30 |
ENSDART00000146776
|
tspan2b
|
tetraspanin 2b |
| chr12_-_36145252 | 0.29 |
|
|
|
| chr5_-_12789388 | 0.28 |
|
|
|
| chr7_-_51877974 | 0.26 |
ENSDART00000122104
|
CT737190.1
|
ENSDARG00000091514 |
| chr12_-_34615030 | 0.25 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr25_+_2623389 | 0.24 |
ENSDART00000156567
|
neo1b
|
neogenin 1b |
| chr13_+_24266204 | 0.23 |
ENSDART00000101200
ENSDART00000166005 |
zgc:153169
|
zgc:153169 |
| chr19_+_19916372 | 0.23 |
ENSDART00000166755
|
tril
|
TLR4 interactor with leucine-rich repeats |
| chr4_+_274394 | 0.22 |
|
|
|
| chr9_+_7609341 | 0.20 |
|
|
|
| chr6_-_7094523 | 0.20 |
|
|
|
| chr17_-_30822468 | 0.20 |
ENSDART00000139707
|
gdf7
|
growth differentiation factor 7 |
| chr20_+_29685331 | 0.19 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr5_-_51351827 | 0.19 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr12_-_34614903 | 0.17 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr9_+_7477732 | 0.16 |
ENSDART00000061723
|
tmem198a
|
transmembrane protein 198a |
| chr12_-_26291790 | 0.15 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr17_-_8570443 | 0.13 |
ENSDART00000049236
|
ctbp2a
|
C-terminal binding protein 2a |
| chr10_-_20524860 | 0.10 |
|
|
|
| chr14_-_50122491 | 0.08 |
ENSDART00000133811
|
sfxn1
|
sideroflexin 1 |
| chr1_-_9201151 | 0.08 |
ENSDART00000138630
|
zgc:56409
|
zgc:56409 |
| chr5_-_21384509 | 0.07 |
ENSDART00000020725
|
smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr4_-_16356247 | 0.05 |
ENSDART00000079521
|
kera
|
keratocan |
| chr11_+_34659252 | 0.04 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr22_-_20141216 | 0.03 |
ENSDART00000128023
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr25_-_31496617 | 0.00 |
ENSDART00000155106
|
ENSDARG00000097988
|
ENSDARG00000097988 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 1.5 | 4.4 | GO:0051701 | interaction with host(GO:0051701) |
| 1.3 | 3.8 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.8 | 2.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.7 | 2.0 | GO:0030431 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.6 | 2.5 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.6 | 1.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.5 | 1.6 | GO:1902869 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.4 | 3.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.4 | 1.4 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.3 | 2.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.3 | 1.9 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.3 | 2.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 1.8 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 1.0 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.2 | 7.9 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.2 | 4.8 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.2 | 1.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 1.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.2 | 4.5 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.2 | 1.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 4.7 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.2 | 2.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.2 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.4 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.1 | 4.0 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 4.0 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 1.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 0.6 | GO:0007100 | mitotic centrosome separation(GO:0007100) positive regulation of centrosome cycle(GO:0046607) centrosome separation(GO:0051299) |
| 0.1 | 2.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.9 | GO:0021986 | habenula development(GO:0021986) |
| 0.1 | 1.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 2.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 5.7 | GO:0021782 | glial cell development(GO:0021782) |
| 0.1 | 2.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 1.4 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 2.7 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.1 | 1.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 3.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 0.6 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 1.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 5.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 4.9 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 2.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 1.2 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 1.0 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.0 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 1.5 | GO:0048793 | pronephros development(GO:0048793) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.6 | 6.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.5 | 1.4 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.5 | 1.9 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.4 | 1.3 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.3 | 1.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 2.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 4.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 1.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 0.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 2.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 7.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 29.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 4.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.1 | GO:0016324 | apical plasma membrane(GO:0016324) apical part of cell(GO:0045177) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.8 | 2.5 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.7 | 4.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.7 | 3.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.6 | 1.8 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.5 | 2.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.5 | 2.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.5 | 4.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.5 | 6.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.5 | 7.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 3.8 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.4 | 3.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.4 | 2.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.3 | 1.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 2.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 1.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 1.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 2.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.4 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.2 | 2.1 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 1.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 2.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 1.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 1.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 1.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 2.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 1.0 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.4 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 1.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.0 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 1.1 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 3.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 4.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 4.0 | GO:0003682 | chromatin binding(GO:0003682) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 7.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 5.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.3 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.9 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.6 | 6.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.4 | 2.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 1.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 2.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |