Project
DANIO-CODE
Navigation
Downloads
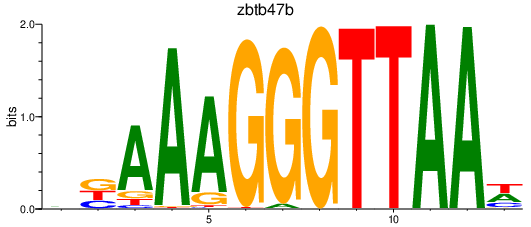
Results for zbtb47b
Z-value: 1.76
Transcription factors associated with zbtb47b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb47b
|
ENSDARG00000079547 | zinc finger and BTB domain containing 47b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb47b | dr10_dc_chr24_-_20496410_20496478 | -0.92 | 3.0e-07 | Click! |
Activity profile of zbtb47b motif
Sorted Z-values of zbtb47b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb47b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23999029 | 10.77 |
ENSDART00000129525
|
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr5_+_37378340 | 6.20 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr20_+_17840364 | 5.10 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr1_+_38834751 | 4.86 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr13_-_33696425 | 4.26 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr13_-_18506153 | 4.13 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr4_-_16556116 | 4.02 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr7_-_26165200 | 3.53 |
ENSDART00000123395
|
her8a
|
hairy-related 8a |
| chr17_-_35934175 | 3.46 |
ENSDART00000110040
ENSDART00000137525 |
sox11a
AL929378.1
|
SRY (sex determining region Y)-box 11a ENSDARG00000092907 |
| chr11_-_44724371 | 3.35 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr18_+_29424354 | 3.32 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr4_+_29855479 | 3.02 |
|
|
|
| chr13_-_34667689 | 2.98 |
ENSDART00000025719
|
ism1
|
isthmin 1 |
| chr14_-_17258072 | 2.97 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr7_-_26035308 | 2.95 |
ENSDART00000131906
|
zgc:77439
|
zgc:77439 |
| chr25_+_31769985 | 2.89 |
|
|
|
| chr4_+_18974767 | 2.86 |
ENSDART00000066973
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr24_-_1155215 | 2.84 |
ENSDART00000177356
|
itgb1a
|
integrin, beta 1a |
| chr4_-_18606513 | 2.83 |
ENSDART00000049061
|
cdkn1ba
|
cyclin-dependent kinase inhibitor 1Ba |
| chr14_+_20632593 | 2.79 |
ENSDART00000166366
ENSDART00000106198 |
zgc:66433
|
zgc:66433 |
| chr7_+_58397256 | 2.72 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr22_-_5652624 | 2.71 |
ENSDART00000127688
ENSDART00000012686 |
dnase1l4.1
|
deoxyribonuclease 1 like 4, tandem duplicate 1 |
| chr25_+_30827761 | 2.61 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr4_-_8610868 | 2.60 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr14_-_17257773 | 2.59 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr10_+_4987494 | 2.52 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr22_-_36915567 | 2.50 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr19_+_7656475 | 2.48 |
ENSDART00000134271
|
s100u
|
S100 calcium binding protein U |
| chr4_-_32820181 | 2.45 |
|
|
|
| chr20_-_22036656 | 2.42 |
ENSDART00000152290
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr13_+_371940 | 2.42 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr19_-_16126342 | 2.40 |
|
|
|
| chr6_+_43018443 | 2.39 |
ENSDART00000064888
|
tcta
|
T-cell leukemia translocation altered gene |
| chr12_+_25006393 | 2.38 |
ENSDART00000050070
|
pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr13_+_7109810 | 2.37 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr21_+_17731439 | 2.34 |
ENSDART00000124173
|
rxraa
|
retinoid X receptor, alpha a |
| chr9_-_42928777 | 2.34 |
ENSDART00000136728
|
fkbp7
|
FK506 binding protein 7 |
| chr9_-_44493074 | 2.32 |
ENSDART00000167685
|
neurod1
|
neuronal differentiation 1 |
| chr7_+_40183670 | 2.28 |
ENSDART00000173916
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr4_+_7406743 | 2.28 |
ENSDART00000168327
|
cald1a
|
caldesmon 1a |
| chr13_-_34667430 | 2.27 |
ENSDART00000025719
|
ism1
|
isthmin 1 |
| chr9_+_16437408 | 2.27 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr7_+_36267647 | 2.25 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr12_+_40919018 | 2.23 |
ENSDART00000176047
|
BX072562.1
|
ENSDARG00000108020 |
| chr19_+_7656513 | 2.23 |
ENSDART00000134271
|
s100u
|
S100 calcium binding protein U |
| chr11_+_3940085 | 2.20 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr19_-_47994946 | 2.19 |
ENSDART00000114549
|
CU695215.1
|
ENSDARG00000076126 |
| chr14_-_49951676 | 2.10 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr23_+_42350554 | 2.10 |
ENSDART00000163730
|
cyp2aa6
|
cytochrome P450, family 2, subfamily AA, polypeptide 6 |
| chr7_+_19300351 | 2.07 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr3_-_55399331 | 2.07 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr20_-_22036622 | 2.04 |
ENSDART00000152290
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr20_+_52778914 | 2.03 |
ENSDART00000138641
|
si:dkey-235d18.5
|
si:dkey-235d18.5 |
| chr10_+_42530040 | 2.02 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr14_-_45883839 | 2.00 |
|
|
|
| chr21_+_20734431 | 1.98 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr21_+_30757831 | 1.97 |
ENSDART00000139486
|
ENSDARG00000030006
|
ENSDARG00000030006 |
| chr4_+_2215426 | 1.95 |
ENSDART00000168370
|
frs2a
|
fibroblast growth factor receptor substrate 2a |
| chr1_+_33563354 | 1.93 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr11_-_3275255 | 1.90 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr4_-_52850069 | 1.85 |
ENSDART00000169714
|
CR388413.6
|
ENSDARG00000108204 |
| chr24_-_39884167 | 1.83 |
ENSDART00000087441
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr4_-_49071258 | 1.82 |
ENSDART00000162560
|
BX855613.1
|
ENSDARG00000098743 |
| chr7_+_20272091 | 1.81 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr8_+_19516856 | 1.80 |
ENSDART00000123104
|
notch2
|
notch 2 |
| chr7_+_15623852 | 1.72 |
ENSDART00000161608
|
pax6b
|
paired box 6b |
| chr24_+_9231693 | 1.71 |
ENSDART00000082422
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr14_+_34626233 | 1.69 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr3_-_45836485 | 1.67 |
|
|
|
| chr24_+_36746202 | 1.66 |
ENSDART00000159017
|
si:ch73-334d15.4
|
si:ch73-334d15.4 |
| chr23_+_42488817 | 1.65 |
ENSDART00000171459
|
cyp2aa2
|
y cytochrome P450, family 2, subfamily AA, polypeptide 2 |
| chr7_+_19857885 | 1.62 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr4_-_24396003 | 1.60 |
|
|
|
| chr7_+_40183905 | 1.60 |
ENSDART00000173916
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr19_-_9192541 | 1.59 |
|
|
|
| chr22_-_10430130 | 1.57 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr9_-_12726136 | 1.56 |
|
|
|
| chr11_-_41241693 | 1.56 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr4_-_29059042 | 1.56 |
ENSDART00000169971
|
CR387997.1
|
ENSDARG00000099486 |
| chr13_+_22350043 | 1.56 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr3_+_48362748 | 1.54 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr20_-_21031504 | 1.53 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr4_-_46037928 | 1.49 |
|
|
|
| chr4_+_149696 | 1.49 |
ENSDART00000161055
|
dusp16
|
dual specificity phosphatase 16 |
| chr2_+_26523457 | 1.49 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr4_+_28860191 | 1.48 |
ENSDART00000110358
ENSDART00000162069 |
znf1059
|
zinc finger protein 1059 |
| chr19_-_22914312 | 1.48 |
|
|
|
| chr19_-_22914516 | 1.47 |
|
|
|
| chr23_-_15574072 | 1.47 |
ENSDART00000035865
ENSDART00000104904 |
sulf2b
|
sulfatase 2b |
| chr13_+_1051015 | 1.46 |
ENSDART00000033528
|
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr12_-_9256949 | 1.43 |
ENSDART00000003805
|
pth1rb
|
parathyroid hormone 1 receptor b |
| chr4_+_58242614 | 1.42 |
|
|
|
| chr14_+_4489377 | 1.41 |
ENSDART00000041468
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr7_+_19857748 | 1.41 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr7_-_40713381 | 1.41 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr19_+_40537428 | 1.41 |
|
|
|
| chr3_+_34064081 | 1.40 |
|
|
|
| chr10_+_25407847 | 1.40 |
ENSDART00000047541
|
bach1b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
| chr6_+_9192009 | 1.40 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr5_-_31708933 | 1.38 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr17_+_48999061 | 1.35 |
ENSDART00000177390
|
tiam2a
|
T-cell lymphoma invasion and metastasis 2a |
| chr17_+_12544451 | 1.35 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr4_+_30012767 | 1.34 |
ENSDART00000150383
|
znf1128
|
zinc finger protein 1128 |
| KN149695v1_+_67009 | 1.33 |
ENSDART00000178024
|
ENSDARG00000107779
|
ENSDARG00000107779 |
| chr23_+_42370612 | 1.33 |
ENSDART00000161812
|
cyp2aa9
|
cytochrome P450, family 2, subfamily AA, polypeptide 9 |
| chr12_+_48761772 | 1.33 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr4_-_16356071 | 1.31 |
ENSDART00000079521
|
kera
|
keratocan |
| chr5_+_68943914 | 1.29 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr8_+_1760968 | 1.28 |
ENSDART00000021820
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr4_+_29577554 | 1.27 |
|
|
|
| chr4_+_51730449 | 1.27 |
|
|
|
| chr1_+_8320586 | 1.26 |
ENSDART00000006211
|
prkcba
|
protein kinase C, beta a |
| chr7_+_39116005 | 1.25 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr3_-_55399433 | 1.25 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr22_+_20110402 | 1.25 |
ENSDART00000110200
|
eef2a.1
|
eukaryotic translation elongation factor 2a, tandem duplicate 1 |
| chr12_-_7902815 | 1.24 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr6_+_44817077 | 1.24 |
ENSDART00000169713
|
chl1b
|
cell adhesion molecule L1-like b |
| chr13_+_19115 | 1.23 |
ENSDART00000176574
|
CABZ01089797.1
|
ENSDARG00000107538 |
| chr4_-_38423096 | 1.23 |
|
|
|
| chr15_-_15513719 | 1.23 |
ENSDART00000101918
|
proca1
|
protein interacting with cyclin A1 |
| chr2_-_23431625 | 1.22 |
ENSDART00000157318
|
BX548024.1
|
ENSDARG00000097593 |
| chr6_-_39491659 | 1.21 |
ENSDART00000151299
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr22_-_36915720 | 1.21 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr7_-_26035541 | 1.19 |
ENSDART00000057288
|
zgc:77439
|
zgc:77439 |
| chr22_-_15930756 | 1.19 |
ENSDART00000080047
|
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr4_+_67053031 | 1.19 |
ENSDART00000158019
|
CR759907.2
|
ENSDARG00000099855 |
| chr21_+_40082890 | 1.18 |
ENSDART00000100166
|
serpinf1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr4_+_43810061 | 1.18 |
ENSDART00000157214
|
si:dkey-7j22.2
|
si:dkey-7j22.2 |
| chr11_-_23206021 | 1.16 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| KN149696v1_+_361745 | 1.16 |
ENSDART00000159019
|
CABZ01074982.1
|
ENSDARG00000099884 |
| chr21_+_22386643 | 1.15 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr23_-_45486779 | 1.14 |
ENSDART00000166883
|
CU682355.1
|
ENSDARG00000102304 |
| chr14_+_51850213 | 1.14 |
|
|
|
| chr4_-_11752109 | 1.14 |
ENSDART00000102301
|
podxl
|
podocalyxin-like |
| chr2_-_57133471 | 1.14 |
|
|
|
| chr25_+_17148124 | 1.14 |
ENSDART00000050379
|
kdm7ab
|
lysine (K)-specific demethylase 7Ab |
| chr10_-_10374032 | 1.12 |
ENSDART00000134929
|
BX323065.1
|
ENSDARG00000095365 |
| chr10_-_21096635 | 1.12 |
ENSDART00000091004
|
pcdh1a
|
protocadherin 1a |
| chr7_+_37105282 | 1.12 |
|
|
|
| chr4_-_29814642 | 1.11 |
ENSDART00000134990
|
znf1119
|
zinc finger protein 1119 |
| chr23_+_23094125 | 1.09 |
ENSDART00000146415
ENSDART00000146463 |
samd11
|
sterile alpha motif domain containing 11 |
| chr19_+_27895343 | 1.09 |
ENSDART00000049368
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr10_-_1690605 | 1.09 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr15_-_794181 | 1.08 |
ENSDART00000157492
|
znf1011
|
zinc finger protein 1011 |
| chr6_-_43679553 | 1.08 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr14_-_7772 | 1.06 |
ENSDART00000054822
|
nkx3.2
|
NK3 homeobox 2 |
| chr21_-_45573799 | 1.06 |
ENSDART00000158489
|
zgc:77058
|
zgc:77058 |
| chr18_+_14627030 | 1.06 |
ENSDART00000080788
|
wfdc1
|
WAP four-disulfide core domain 1 |
| chr4_+_57490141 | 1.05 |
|
|
|
| chr12_+_23717554 | 1.05 |
ENSDART00000152997
|
svila
|
supervillin a |
| chr21_+_30758036 | 1.04 |
ENSDART00000139486
|
ENSDARG00000030006
|
ENSDARG00000030006 |
| chr3_+_45375639 | 1.03 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr20_-_26638025 | 1.03 |
ENSDART00000061926
|
stx11b.2
|
syntaxin 11b, tandem duplicate 2 |
| chr19_+_19939410 | 1.03 |
|
|
|
| chr25_-_28948049 | 1.01 |
|
|
|
| chr7_+_28896401 | 1.01 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr15_+_28370102 | 1.00 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr22_-_37676744 | 1.00 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr20_+_28958586 | 0.99 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr4_-_44504315 | 0.99 |
ENSDART00000177682
|
znf1046
|
zinc finger protein 1046 |
| chr10_+_44195410 | 0.99 |
ENSDART00000046172
|
cryba4
|
crystallin, beta A4 |
| chr18_-_1084561 | 0.99 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr17_+_48999102 | 0.98 |
ENSDART00000177390
|
tiam2a
|
T-cell lymphoma invasion and metastasis 2a |
| chr13_+_13799335 | 0.98 |
ENSDART00000169995
ENSDART00000165619 |
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr4_+_57648974 | 0.98 |
ENSDART00000108499
|
znf1068
|
zinc finger protein 1068 |
| chr16_+_5302404 | 0.98 |
ENSDART00000145368
|
soga3a
|
SOGA family member 3a |
| chr7_-_30289159 | 0.97 |
ENSDART00000142818
|
sltm
|
SAFB-like, transcription modulator |
| chr19_-_33341287 | 0.96 |
ENSDART00000050750
|
rrm2b
|
ribonucleotide reductase M2 b |
| chr18_-_14868444 | 0.96 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr17_-_1599376 | 0.95 |
ENSDART00000034216
ENSDART00000158837 |
dync1h1
|
dynein, cytoplasmic 1, heavy chain 1 |
| chr4_+_52851409 | 0.95 |
|
|
|
| chr22_-_37676556 | 0.95 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr24_+_9231768 | 0.95 |
ENSDART00000082422
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr4_-_55040981 | 0.94 |
ENSDART00000158368
|
BX957361.1
|
ENSDARG00000101664 |
| chr3_-_45040094 | 0.94 |
|
|
|
| chr20_-_34898276 | 0.94 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr3_-_12818954 | 0.93 |
ENSDART00000158747
ENSDART00000158815 |
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr16_-_27743294 | 0.93 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr3_+_49166063 | 0.93 |
ENSDART00000156347
|
epn3a
|
epsin 3a |
| chr18_+_17552191 | 0.92 |
|
|
|
| chr6_-_49674729 | 0.92 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr15_-_4500676 | 0.91 |
|
|
|
| chr6_+_53349584 | 0.91 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr6_+_29096734 | 0.91 |
|
|
|
| chr4_-_38406131 | 0.89 |
|
|
|
| chr4_-_35766769 | 0.89 |
ENSDART00000163144
|
zgc:174653
|
zgc:174653 |
| chr5_+_27298136 | 0.89 |
ENSDART00000098604
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr19_+_7656412 | 0.88 |
ENSDART00000134271
|
s100u
|
S100 calcium binding protein U |
| chr6_-_2501966 | 0.88 |
ENSDART00000155109
|
hyi
|
hydroxypyruvate isomerase |
| chr4_-_28651054 | 0.88 |
ENSDART00000167212
|
CABZ01050663.1
|
ENSDARG00000104584 |
| chr20_-_19466153 | 0.88 |
ENSDART00000161065
|
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr12_+_34615991 | 0.87 |
|
|
|
| chr4_+_25868802 | 0.87 |
|
|
|
| chr2_+_29992879 | 0.87 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr16_+_6002342 | 0.86 |
ENSDART00000060519
|
ulk4
|
unc-51 like kinase 4 |
| chr21_+_30757998 | 0.85 |
ENSDART00000139486
|
ENSDARG00000030006
|
ENSDARG00000030006 |
| chr4_+_41633430 | 0.85 |
ENSDART00000162193
|
si:ch211-142b24.2
|
si:ch211-142b24.2 |
| chr14_+_51692872 | 0.84 |
ENSDART00000163856
|
noa1
|
nitric oxide associated 1 |
| chr23_+_23093658 | 0.84 |
ENSDART00000146415
ENSDART00000146463 |
samd11
|
sterile alpha motif domain containing 11 |
| chr4_-_11751943 | 0.84 |
ENSDART00000102301
|
podxl
|
podocalyxin-like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0034434 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) reverse cholesterol transport(GO:0043691) positive regulation of steroid metabolic process(GO:0045940) |
| 1.7 | 5.1 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 1.6 | 4.9 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 1.1 | 3.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.8 | 2.3 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.7 | 2.0 | GO:0033630 | regulation of cell adhesion mediated by integrin(GO:0033628) positive regulation of cell adhesion mediated by integrin(GO:0033630) cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.6 | 4.2 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.6 | 2.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.6 | 5.6 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.5 | 2.0 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.5 | 2.0 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.5 | 1.5 | GO:0071947 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.5 | 5.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.4 | 1.3 | GO:1902855 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.4 | 1.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.4 | 2.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.4 | 2.8 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 1.7 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.3 | 4.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.3 | 3.3 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.3 | 0.3 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.2 | 2.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 0.7 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.2 | 1.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 1.4 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) |
| 0.2 | 3.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.2 | 1.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 1.8 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.2 | 2.4 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.2 | 1.5 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 1.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 3.0 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.2 | 2.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 2.9 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.2 | 1.2 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 2.6 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.2 | 1.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 1.4 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 0.9 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 1.9 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 2.9 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 3.0 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 2.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 1.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.8 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.2 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.1 | 1.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 4.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 0.3 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 0.5 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 1.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 10.9 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.1 | 0.3 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.1 | 1.0 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.8 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 0.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 2.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 2.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.7 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 0.5 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 0.6 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.1 | 0.2 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 1.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 1.0 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 3.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 1.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 4.0 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.1 | 1.0 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 1.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 3.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 2.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 4.8 | GO:0010001 | glial cell differentiation(GO:0010001) |
| 0.0 | 2.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.6 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.8 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 2.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.0 | 1.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.3 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.4 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 1.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.9 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 2.0 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.5 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 2.7 | GO:0060026 | convergent extension(GO:0060026) |
| 0.0 | 1.1 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 1.0 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.1 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 2.7 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 2.4 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.3 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.5 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.2 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.2 | GO:0042627 | chylomicron(GO:0042627) |
| 1.1 | 3.2 | GO:0098594 | mucin granule(GO:0098594) |
| 0.4 | 5.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.3 | 1.7 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.3 | 2.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.2 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 4.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 3.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.8 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 6.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.6 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 3.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.8 | GO:0009295 | nucleoid(GO:0009295) ribonuclease P complex(GO:0030677) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 4.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 0.9 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 0.2 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 1.2 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 1.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 6.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.0 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 2.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.0 | 0.7 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 21.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.0 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.4 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.2 | GO:0071814 | lipoprotein particle binding(GO:0071813) protein-lipid complex binding(GO:0071814) |
| 0.9 | 2.8 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.8 | 4.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.7 | 4.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.5 | 3.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 3.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.5 | 2.0 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.5 | 2.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.5 | 2.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 1.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.4 | 5.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.4 | 6.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 2.9 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 1.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 1.3 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.3 | 2.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.0 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.2 | 3.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.2 | 2.8 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.8 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 1.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 2.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 5.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 2.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 5.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 3.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.3 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 4.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 1.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 3.6 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.1 | 3.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 2.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 1.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.6 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 1.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 4.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 2.2 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 2.7 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen(GO:0016705) |
| 0.0 | 0.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 9.0 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 8.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 4.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 24.0 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 1.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.3 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0051427 | hormone receptor binding(GO:0051427) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 2.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 0.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 5.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.4 | 1.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 2.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 5.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 5.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 4.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.0 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.1 | 1.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |