Project
DANIO-CODE
Navigation
Downloads
Results for zbtb7b
Z-value: 0.72
Transcription factors associated with zbtb7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb7b
|
ENSDARG00000021891 | zinc finger and BTB domain containing 7B |
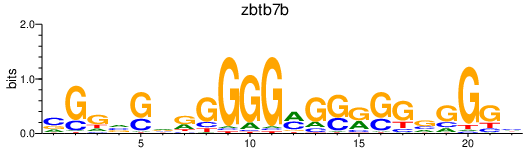
Activity profile of zbtb7b motif
Sorted Z-values of zbtb7b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb7b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_23965038 | 2.13 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr10_-_25695574 | 1.30 |
ENSDART00000110751
|
tiam1a
|
T-cell lymphoma invasion and metastasis 1a |
| chr14_-_6901415 | 1.24 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr7_-_54407461 | 1.14 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr14_-_6901783 | 1.10 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr16_+_55167489 | 1.08 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr3_+_22412059 | 1.02 |
|
|
|
| chr11_+_44286283 | 0.98 |
ENSDART00000168209
|
nid1b
|
nidogen 1b |
| chr8_-_52952226 | 0.97 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr3_-_23513177 | 0.91 |
ENSDART00000078425
|
eve1
|
even-skipped-like1 |
| chr1_-_50831155 | 0.89 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr7_+_19300351 | 0.88 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr20_-_21773202 | 0.88 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr13_+_22173410 | 0.84 |
ENSDART00000173405
|
usp54a
|
ubiquitin specific peptidase 54a |
| chr11_-_29891067 | 0.80 |
ENSDART00000172106
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr23_+_36832325 | 0.79 |
|
|
|
| chr10_-_5016621 | 0.76 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr18_-_26876598 | 0.75 |
|
|
|
| chr20_-_25732091 | 0.73 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr8_-_52952141 | 0.73 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr15_+_42440802 | 0.70 |
ENSDART00000089694
|
tiam1b
|
T-cell lymphoma invasion and metastasis 1b |
| chr17_+_26946957 | 0.63 |
ENSDART00000114215
|
grhl3
|
grainyhead-like transcription factor 3 |
| chr8_+_584353 | 0.63 |
ENSDART00000048498
|
CABZ01049925.1
|
ENSDARG00000079790 |
| chr1_-_51862897 | 0.62 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| KN150256v1_-_9467 | 0.61 |
|
|
|
| chr20_-_21773327 | 0.61 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr17_-_8155740 | 0.61 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr15_+_42440952 | 0.61 |
ENSDART00000089694
|
tiam1b
|
T-cell lymphoma invasion and metastasis 1b |
| chr1_-_46015367 | 0.61 |
ENSDART00000074519
|
kpna3
|
karyopherin alpha 3 (importin alpha 4) |
| chr25_-_34638406 | 0.60 |
ENSDART00000123853
|
hist1h4l
|
histone 1, H4, like |
| chr22_-_21021645 | 0.51 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr11_-_471166 | 0.50 |
ENSDART00000154888
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr4_-_76594677 | 0.48 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr8_-_7425677 | 0.45 |
ENSDART00000149671
|
hdac6
|
histone deacetylase 6 |
| chr3_-_25239180 | 0.45 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr16_-_31476944 | 0.43 |
ENSDART00000167350
|
zgc:194210
|
zgc:194210 |
| chr7_+_16610457 | 0.42 |
|
|
|
| chr7_-_56533829 | 0.41 |
ENSDART00000010322
|
senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr4_-_22751641 | 0.40 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr10_-_20524586 | 0.40 |
|
|
|
| chr19_-_47994946 | 0.39 |
ENSDART00000114549
|
CU695215.1
|
ENSDARG00000076126 |
| chr9_-_264078 | 0.39 |
ENSDART00000166231
|
pcbp2
|
poly(rC) binding protein 2 |
| chr19_-_5142310 | 0.38 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr5_+_36299911 | 0.36 |
ENSDART00000142960
|
hnrpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr4_-_57935184 | 0.36 |
|
|
|
| KN149861v1_-_8998 | 0.34 |
|
|
|
| KN150421v1_+_14181 | 0.34 |
|
|
|
| chr22_-_95158 | 0.33 |
|
|
|
| chr10_-_45134642 | 0.33 |
ENSDART00000166528
|
purbb
|
purine-rich element binding protein Bb |
| chr19_-_22914516 | 0.33 |
|
|
|
| chr3_+_22411884 | 0.31 |
|
|
|
| chr16_+_53178713 | 0.31 |
ENSDART00000174634
|
CABZ01062546.1
|
ENSDARG00000108858 |
| chr10_-_44675914 | 0.30 |
|
|
|
| chr10_+_44210299 | 0.30 |
ENSDART00000164610
|
adrbk2
|
adrenergic, beta, receptor kinase 2 |
| chr25_-_36760720 | 0.30 |
ENSDART00000111862
|
ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr6_-_43679553 | 0.29 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr15_+_6789662 | 0.28 |
ENSDART00000013278
|
sirt2
|
sirtuin 2 (silent mating type information regulation 2, homolog) 2 (S. cerevisiae) |
| chr12_-_954375 | 0.27 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr20_-_29580624 | 0.27 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr23_+_44951868 | 0.25 |
|
|
|
| chr6_-_43094573 | 0.25 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr23_-_9928561 | 0.25 |
|
|
|
| chr13_-_25067585 | 0.24 |
ENSDART00000159585
|
adka
|
adenosine kinase a |
| chr4_-_12930797 | 0.24 |
ENSDART00000108552
|
lemd3
|
LEM domain containing 3 |
| chr12_+_16832804 | 0.24 |
|
|
|
| chr3_-_20832321 | 0.23 |
ENSDART00000055722
|
slc35b1
|
solute carrier family 35, member B1 |
| chr17_+_8166308 | 0.23 |
ENSDART00000172443
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr16_+_25845432 | 0.23 |
|
|
|
| chr23_+_36002332 | 0.23 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr19_-_22914312 | 0.23 |
|
|
|
| chr4_+_11312432 | 0.22 |
ENSDART00000051792
|
sema3aa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Aa |
| chr8_+_17479651 | 0.22 |
|
|
|
| chr21_-_11554253 | 0.22 |
ENSDART00000133443
|
cast
|
calpastatin |
| chr13_-_35781888 | 0.22 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr10_+_22065010 | 0.22 |
ENSDART00000133304
|
npm1a
|
nucleophosmin 1a (nucleolar phosphoprotein B23, numatrin) |
| chr19_-_17039269 | 0.21 |
|
|
|
| chr7_+_39130411 | 0.21 |
ENSDART00000144750
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr19_-_22914419 | 0.21 |
|
|
|
| chr25_-_28691412 | 0.21 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr21_-_45833636 | 0.21 |
ENSDART00000161716
|
larp1
|
La ribonucleoprotein domain family, member 1 |
| chr22_+_9493668 | 0.20 |
ENSDART00000143953
|
strip1
|
striatin interacting protein 1 |
| chr12_-_36138709 | 0.20 |
ENSDART00000130985
|
st6galnac
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase |
| KN150256v1_-_9560 | 0.20 |
|
|
|
| chr23_+_22408855 | 0.20 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr14_+_46609215 | 0.19 |
ENSDART00000110429
|
zgc:194285
|
zgc:194285 |
| chr20_+_35155682 | 0.19 |
ENSDART00000165899
ENSDART00000122696 |
hnrnpub
|
heterogeneous nuclear ribonucleoprotein Ub |
| chr14_+_30388744 | 0.18 |
|
|
|
| chr22_+_10669147 | 0.18 |
ENSDART00000081228
|
abhd14a
|
abhydrolase domain containing 14A |
| chr19_-_2486568 | 0.18 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr20_+_35156202 | 0.18 |
ENSDART00000165899
ENSDART00000122696 |
hnrnpub
|
heterogeneous nuclear ribonucleoprotein Ub |
| chr22_-_21021984 | 0.18 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr10_-_5016472 | 0.18 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr10_+_12559022 | 0.17 |
ENSDART00000159853
|
FQ311890.1
|
ENSDARG00000102794 |
| chr16_-_9108885 | 0.17 |
ENSDART00000153785
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr4_-_1809690 | 0.17 |
|
|
|
| chr17_+_26946875 | 0.17 |
ENSDART00000114215
|
grhl3
|
grainyhead-like transcription factor 3 |
| chr18_+_22185526 | 0.17 |
ENSDART00000089549
|
fam65a
|
family with sequence similarity 65, member A |
| chr2_+_43619752 | 0.16 |
ENSDART00000142078
ENSDART00000098265 ENSDART00000098267 |
nrp1b
|
neuropilin 1b |
| chr5_-_13145749 | 0.16 |
ENSDART00000166957
|
purba
|
purine-rich element binding protein Ba |
| chr3_+_14238188 | 0.16 |
ENSDART00000165452
ENSDART00000171726 |
tmem56b
|
transmembrane protein 56b |
| chr3_+_15645543 | 0.16 |
|
|
|
| chr7_-_57030751 | 0.16 |
|
|
|
| chr13_-_50053533 | 0.16 |
ENSDART00000098209
|
sirt1
|
sirtuin 1 |
| chr19_-_12226198 | 0.16 |
ENSDART00000032474
ENSDART00000151056 ENSDART00000167360 |
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr24_+_26944182 | 0.15 |
ENSDART00000089541
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| KN150640v1_+_5728 | 0.15 |
|
|
|
| chr16_+_11138879 | 0.15 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr11_-_14943957 | 0.14 |
ENSDART00000162938
|
hug
|
HuG |
| chr15_+_31036717 | 0.14 |
ENSDART00000129474
|
ogmb
|
oligodendrocyte myelin glycoprotein b |
| chr16_+_29716279 | 0.14 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr3_-_23513293 | 0.14 |
ENSDART00000140264
|
eve1
|
even-skipped-like1 |
| chr17_+_52438099 | 0.14 |
ENSDART00000135246
|
dlst
|
dihydrolipoamide S-succinyltransferase |
| chr20_+_35156247 | 0.14 |
ENSDART00000165899
ENSDART00000122696 |
hnrnpub
|
heterogeneous nuclear ribonucleoprotein Ub |
| chr1_+_40795366 | 0.13 |
ENSDART00000146310
|
dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr5_+_69401770 | 0.13 |
|
|
|
| chr20_-_54645903 | 0.13 |
ENSDART00000038745
|
yy1b
|
YY1 transcription factor b |
| chr1_-_22144014 | 0.13 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr13_-_35782030 | 0.13 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr15_+_42441012 | 0.12 |
ENSDART00000089694
|
tiam1b
|
T-cell lymphoma invasion and metastasis 1b |
| chr2_-_44893769 | 0.12 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr23_+_39454172 | 0.12 |
ENSDART00000149819
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr16_+_54729975 | 0.11 |
|
|
|
| chr14_+_23986703 | 0.11 |
ENSDART00000079164
|
klhl3
|
kelch-like family member 3 |
| chr7_-_38205976 | 0.10 |
ENSDART00000084355
|
zgc:165481
|
zgc:165481 |
| chr20_+_35156072 | 0.10 |
ENSDART00000165899
ENSDART00000122696 |
hnrnpub
|
heterogeneous nuclear ribonucleoprotein Ub |
| chr4_-_76594399 | 0.10 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr16_+_11138924 | 0.10 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr6_-_43094926 | 0.10 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr19_+_1298795 | 0.09 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr17_+_32547862 | 0.09 |
ENSDART00000018423
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr3_-_4590689 | 0.09 |
ENSDART00000163052
ENSDART00000164569 |
ENSDARG00000098540
|
ENSDARG00000098540 |
| chr21_-_45833527 | 0.08 |
ENSDART00000161716
|
larp1
|
La ribonucleoprotein domain family, member 1 |
| chr20_-_54645287 | 0.08 |
ENSDART00000153389
|
yy1b
|
YY1 transcription factor b |
| chr23_-_6442213 | 0.08 |
ENSDART00000112667
|
ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr7_+_36281268 | 0.08 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr17_+_52736192 | 0.07 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr6_-_43679476 | 0.07 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr22_-_21021866 | 0.07 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr6_+_52212513 | 0.05 |
ENSDART00000025940
|
ywhaba
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a |
| chr25_+_4806680 | 0.05 |
|
|
|
| chr13_+_49154065 | 0.05 |
|
|
|
| chr8_-_44941251 | 0.05 |
|
|
|
| chr18_+_30391910 | 0.05 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
| chr1_+_28936288 | 0.05 |
|
|
|
| chr7_-_69399273 | 0.05 |
ENSDART00000159823
ENSDART00000075178 ENSDART00000168942 ENSDART00000126739 |
tspan5a
|
tetraspanin 5a |
| chr13_+_31586024 | 0.04 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| KN149932v1_+_27584 | 0.04 |
|
|
|
| chr2_+_24718945 | 0.04 |
ENSDART00000133818
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr15_-_18087085 | 0.04 |
ENSDART00000155194
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr13_+_45445475 | 0.04 |
|
|
|
| chr7_+_19300487 | 0.04 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr5_-_68270398 | 0.04 |
ENSDART00000161561
|
ENSDARG00000059093
|
ENSDARG00000059093 |
| chr20_-_51915872 | 0.03 |
ENSDART00000004092
|
brox
|
BRO1 domain and CAAX motif containing |
| chr5_-_36647498 | 0.03 |
ENSDART00000112312
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr25_+_25369539 | 0.03 |
ENSDART00000150537
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr6_-_10574082 | 0.03 |
ENSDART00000151661
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr13_+_291022 | 0.03 |
ENSDART00000017854
|
lgi1a
|
leucine-rich, glioma inactivated 1a |
| KN150411v1_-_12184 | 0.03 |
|
|
|
| chr13_+_290930 | 0.03 |
ENSDART00000017854
|
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr21_+_19889374 | 0.03 |
ENSDART00000111694
|
tnksa
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase a |
| chr25_+_33364224 | 0.02 |
ENSDART00000121449
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr6_+_36843778 | 0.02 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr21_+_15505169 | 0.02 |
ENSDART00000011318
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr23_+_22645604 | 0.02 |
ENSDART00000177273
|
CR388207.2
|
ENSDARG00000108862 |
| chr5_+_2486144 | 0.02 |
ENSDART00000160815
|
ENSDARG00000102078
|
ENSDARG00000102078 |
| chr21_-_25379720 | 0.01 |
ENSDART00000114081
ENSDART00000161629 |
sms
|
spermine synthase |
| chr18_+_30392138 | 0.01 |
ENSDART00000165450
|
gse1
|
Gse1 coiled-coil protein |
| chr10_+_34372283 | 0.01 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr11_+_44916426 | 0.01 |
|
|
|
| chr19_-_5142538 | 0.01 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr20_-_54646114 | 0.01 |
ENSDART00000038745
|
yy1b
|
YY1 transcription factor b |
| chr20_+_33972327 | 0.01 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr19_+_14490375 | 0.01 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr1_-_13767901 | 0.01 |
|
|
|
| chr3_+_52951742 | 0.01 |
|
|
|
| chr13_-_49153890 | 0.01 |
ENSDART00000136991
|
irf2bp2a
|
interferon regulatory factor 2 binding protein 2a |
| chr10_-_1645 | 0.00 |
|
|
|
| chr2_+_31492528 | 0.00 |
ENSDART00000130722
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr22_+_9493775 | 0.00 |
ENSDART00000143953
|
strip1
|
striatin interacting protein 1 |
| chr3_-_60930025 | 0.00 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic G1 DNA damage checkpoint(GO:0031571) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 | 0.9 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.6 | GO:1902425 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) regulation of mitotic attachment of spindle microtubules to kinetochore(GO:1902423) positive regulation of attachment of mitotic spindle microtubules to kinetochore(GO:1902425) RNA localization to chromatin(GO:1990280) |
| 0.1 | 1.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.3 | GO:2000378 | regulation of DNA binding(GO:0051101) regulation of oligodendrocyte progenitor proliferation(GO:0070445) negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.3 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.1 | 1.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 1.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 2.7 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 1.1 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.2 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.1 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.6 | GO:0003128 | heart field specification(GO:0003128) |
| 0.0 | 0.2 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 1.7 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.2 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.2 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 1.0 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.6 | GO:0000803 | sex chromosome(GO:0000803) inactive sex chromosome(GO:0098577) |
| 0.1 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.4 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 0.7 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.6 | GO:0017130 | TFIIH-class transcription factor binding(GO:0001097) poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0005460 | UDP-glucose transmembrane transporter activity(GO:0005460) |
| 0.0 | 0.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 2.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |