Project
DANIO-CODE
Navigation
Downloads
Results for zbtb7c
Z-value: 0.40
Transcription factors associated with zbtb7c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb7c
|
ENSDARG00000100985 | zinc finger and BTB domain containing 7C |
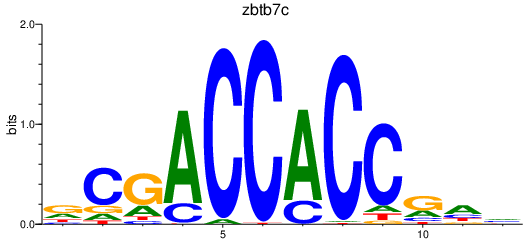
Activity profile of zbtb7c motif
Sorted Z-values of zbtb7c motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb7c
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_3323004 | 0.52 |
|
|
|
| chr5_+_22006860 | 0.51 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr5_+_30880165 | 0.39 |
ENSDART00000098197
|
ENSDARG00000035471
|
ENSDARG00000035471 |
| chr23_+_36209868 | 0.38 |
ENSDART00000131711
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr24_-_13118075 | 0.36 |
|
|
|
| chr23_+_36209604 | 0.35 |
ENSDART00000134607
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr12_-_4597113 | 0.33 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr3_-_39554155 | 0.32 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr10_-_24401876 | 0.28 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr14_+_45893080 | 0.28 |
ENSDART00000173167
|
pdlim7
|
PDZ and LIM domain 7 |
| chr23_+_36209696 | 0.26 |
ENSDART00000134607
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr9_+_56881036 | 0.25 |
|
|
|
| chr23_+_28656263 | 0.24 |
ENSDART00000020296
|
nadl1.2
|
neural adhesion molecule L1.2 |
| chr11_+_34659252 | 0.23 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr23_+_44543892 | 0.22 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr4_+_17290988 | 0.22 |
ENSDART00000067005
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr2_-_38242982 | 0.21 |
ENSDART00000040357
|
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr17_+_34047037 | 0.20 |
ENSDART00000155030
ENSDART00000153942 ENSDART00000167085 ENSDART00000127692 |
gphna
|
gephyrin a |
| chr20_-_35109185 | 0.20 |
ENSDART00000062761
|
cnstb
|
consortin, connexin sorting protein b |
| chr10_+_26554237 | 0.19 |
ENSDART00000134276
|
synj1
|
synaptojanin 1 |
| chr7_+_38690837 | 0.19 |
ENSDART00000173565
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr25_-_12728011 | 0.19 |
ENSDART00000169126
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr14_-_49115338 | 0.19 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr7_-_44332679 | 0.18 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr7_-_28771601 | 0.17 |
ENSDART00000052342
|
thap11
|
THAP domain containing 11 |
| chr3_-_11861502 | 0.17 |
ENSDART00000018159
|
ENSDARG00000011609
|
ENSDARG00000011609 |
| chr17_-_30958965 | 0.16 |
ENSDART00000051697
|
evla
|
Enah/Vasp-like a |
| chr7_+_59773957 | 0.16 |
ENSDART00000051524
|
etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr17_-_21258714 | 0.15 |
ENSDART00000157518
|
hspa12a
|
heat shock protein 12A |
| chr8_-_52105120 | 0.14 |
ENSDART00000108923
ENSDART00000111204 ENSDART00000163457 |
vps13a
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr13_+_51485138 | 0.14 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr8_-_7584890 | 0.14 |
ENSDART00000137975
|
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr20_+_51034244 | 0.13 |
|
|
|
| chr5_-_47519273 | 0.13 |
|
|
|
| chr14_+_45893178 | 0.12 |
ENSDART00000173167
|
pdlim7
|
PDZ and LIM domain 7 |
| chr5_+_22006830 | 0.12 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr3_+_55994886 | 0.12 |
|
|
|
| chr12_-_17370920 | 0.11 |
ENSDART00000130735
|
minpp1b
|
multiple inositol-polyphosphate phosphatase 1b |
| chr17_+_43916865 | 0.11 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr3_-_39554099 | 0.11 |
ENSDART00000145303
|
b9d1
|
B9 protein domain 1 |
| chr23_+_36209223 | 0.11 |
ENSDART00000134607
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr6_+_29716538 | 0.11 |
ENSDART00000164849
|
phb2b
|
prohibitin 2b |
| chr2_+_38243131 | 0.11 |
ENSDART00000141614
|
si:ch211-14a17.11
|
si:ch211-14a17.11 |
| chr3_+_22724101 | 0.11 |
ENSDART00000167127
|
BX322574.1
|
ENSDARG00000098353 |
| chr12_+_17370986 | 0.11 |
|
|
|
| chr14_-_1186603 | 0.10 |
|
|
|
| chr25_-_12712755 | 0.10 |
ENSDART00000162750
|
ca5a
|
carbonic anhydrase Va |
| chr12_+_25509394 | 0.10 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr6_+_40954138 | 0.10 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr11_-_37921642 | 0.10 |
|
|
|
| chr12_-_17370608 | 0.09 |
ENSDART00000012421
ENSDART00000138766 |
minpp1b
|
multiple inositol-polyphosphate phosphatase 1b |
| chr14_-_1186789 | 0.09 |
|
|
|
| chr1_-_51494196 | 0.09 |
ENSDART00000149939
|
rad23aa
|
RAD23 homolog A, nucleotide excision repair protein a |
| chr3_-_11861438 | 0.09 |
ENSDART00000018159
|
ENSDARG00000011609
|
ENSDARG00000011609 |
| chr14_-_1187074 | 0.09 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr16_+_21121428 | 0.08 |
|
|
|
| chr3_+_32279283 | 0.08 |
ENSDART00000061426
|
rras
|
related RAS viral (r-ras) oncogene homolog |
| chr1_+_9999665 | 0.08 |
ENSDART00000054879
|
ENSDARG00000037679
|
ENSDARG00000037679 |
| chr4_-_17735989 | 0.08 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr8_-_18502159 | 0.07 |
ENSDART00000148802
ENSDART00000149081 ENSDART00000148962 |
nexn
|
nexilin (F actin binding protein) |
| chr14_-_46359062 | 0.07 |
ENSDART00000090844
|
zgc:153018
|
zgc:153018 |
| chr16_+_27608672 | 0.07 |
ENSDART00000143708
|
si:ch211-197h24.6
|
si:ch211-197h24.6 |
| chr3_+_57878398 | 0.07 |
ENSDART00000047418
|
notum1a
|
notum pectinacetylesterase homolog 1a (Drosophila) |
| chr9_+_36051871 | 0.07 |
ENSDART00000144141
|
rcan1a
|
regulator of calcineurin 1a |
| chr20_+_37397782 | 0.06 |
ENSDART00000067053
|
vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr21_+_7543440 | 0.06 |
|
|
|
| chr6_-_22088774 | 0.05 |
ENSDART00000168932
|
tefm
|
transcription elongation factor, mitochondrial |
| chr7_-_22685672 | 0.05 |
ENSDART00000101447
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr6_-_22834226 | 0.05 |
ENSDART00000165484
|
jpt1a
|
Jupiter microtubule associated homolog 1a |
| chr6_+_20008821 | 0.05 |
ENSDART00000151205
|
aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr25_+_14153652 | 0.05 |
|
|
|
| chr17_-_24860499 | 0.04 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr7_-_33078758 | 0.04 |
ENSDART00000008785
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr16_+_21122024 | 0.04 |
|
|
|
| chr17_-_42108276 | 0.03 |
ENSDART00000110871
ENSDART00000155484 |
nkx2.4a
|
NK2 homeobox 4a |
| chr7_+_44265618 | 0.03 |
ENSDART00000171690
ENSDART00000161949 |
CT027744.2
|
ENSDARG00000102431 |
| chr1_-_18809429 | 0.03 |
ENSDART00000124260
|
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr21_-_41611442 | 0.02 |
ENSDART00000100039
|
pcyox1l
|
prenylcysteine oxidase 1 like |
| chr4_+_13811932 | 0.02 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr7_-_22685631 | 0.02 |
ENSDART00000122113
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr19_+_4940844 | 0.02 |
|
|
|
| chr10_-_14530112 | 0.02 |
ENSDART00000176126
|
galt
|
galactose-1-phosphate uridylyltransferase |
| chr11_+_24662538 | 0.01 |
ENSDART00000148023
|
timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr14_+_26137608 | 0.01 |
ENSDART00000175459
|
gpr137
|
G protein-coupled receptor 137 |
| KN149861v1_-_6661 | 0.01 |
|
|
|
| chr19_-_43037791 | 0.01 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr16_-_19154824 | 0.01 |
ENSDART00000088818
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr20_+_43216682 | 0.00 |
ENSDART00000004842
|
dusp23a
|
dual specificity phosphatase 23a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) |
| 0.1 | 0.2 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.2 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.0 | 0.2 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.2 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.2 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.1 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |