Project
DANIO-CODE
Navigation
Downloads
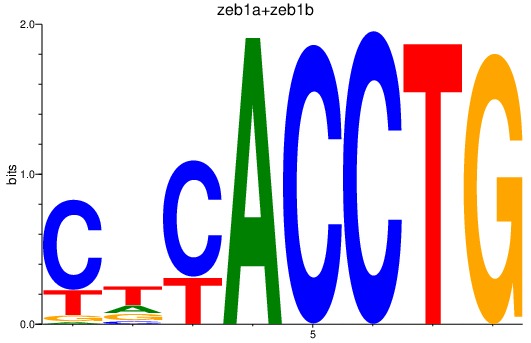
Results for zeb1a+zeb1b
Z-value: 3.11
Transcription factors associated with zeb1a+zeb1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zeb1b
|
ENSDARG00000013207 | zinc finger E-box binding homeobox 1b |
|
zeb1a
|
ENSDARG00000016788 | zinc finger E-box binding homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zeb1b | dr10_dc_chr12_-_26760324_26760394 | -0.93 | 1.7e-07 | Click! |
| zeb1a | dr10_dc_chr2_-_43997672_43997784 | -0.85 | 2.7e-05 | Click! |
Activity profile of zeb1a+zeb1b motif
Sorted Z-values of zeb1a+zeb1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zeb1a+zeb1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_55346967 | 14.17 |
ENSDART00000135304
|
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr21_-_30256477 | 11.62 |
ENSDART00000137193
|
slbp2
|
stem-loop binding protein 2 |
| chr24_+_12689711 | 9.87 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr14_+_22159893 | 7.92 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr5_-_32636372 | 7.79 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr7_-_73890499 | 7.76 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr23_+_540624 | 7.72 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr7_+_58448953 | 7.52 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr21_-_804453 | 7.33 |
|
|
|
| chr14_+_29945327 | 7.29 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr14_+_24543399 | 7.22 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr23_-_18131502 | 7.20 |
ENSDART00000173075
|
zgc:92287
|
zgc:92287 |
| chr16_-_25318413 | 7.05 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr1_-_33717934 | 6.25 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr6_+_135002 | 6.23 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr24_+_5771253 | 6.14 |
ENSDART00000154930
|
si:ch211-157j23.2
|
si:ch211-157j23.2 |
| chr24_+_5782465 | 6.13 |
ENSDART00000178238
|
si:ch211-157j23.3
|
si:ch211-157j23.3 |
| chr1_-_44892852 | 6.05 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr15_+_29092022 | 5.93 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr23_+_2786407 | 5.88 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr13_-_6875290 | 5.87 |
|
|
|
| chr8_-_18637047 | 5.83 |
ENSDART00000172584
ENSDART00000100516 |
stap2b
|
signal transducing adaptor family member 2b |
| chr5_-_67121850 | 5.67 |
ENSDART00000017881
|
eif4e1b
|
eukaryotic translation initiation factor 4E family member 1B |
| chr21_-_3548863 | 5.56 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr12_+_13704712 | 5.55 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr1_-_44892600 | 5.55 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr14_+_14850200 | 5.52 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr9_+_38832959 | 5.50 |
ENSDART00000110651
|
slc12a8
|
solute carrier family 12, member 8 |
| chr14_-_7102535 | 5.50 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr15_-_43283562 | 5.40 |
ENSDART00000110352
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr19_+_15536640 | 5.40 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr24_-_38195383 | 5.29 |
ENSDART00000056381
|
crp2
|
C-reactive protein 2 |
| KN150589v1_-_5209 | 5.28 |
ENSDART00000157761
ENSDART00000157531 |
elovl7b
|
ELOVL fatty acid elongase 7b |
| chr19_+_2942485 | 5.13 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr9_-_28444272 | 5.05 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr24_+_12689887 | 5.00 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr22_-_24964889 | 4.98 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr10_-_7513764 | 4.94 |
ENSDART00000167054
ENSDART00000167706 |
nrg1
|
neuregulin 1 |
| chr10_-_25448769 | 4.92 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr7_+_58449163 | 4.91 |
ENSDART00000167166
|
zgc:56231
|
zgc:56231 |
| chr2_-_50491234 | 4.90 |
ENSDART00000165678
|
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr25_+_7067115 | 4.86 |
ENSDART00000179500
|
BX005076.1
|
ENSDARG00000107609 |
| chr12_+_13053552 | 4.84 |
ENSDART00000124799
|
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr10_-_2915710 | 4.80 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr18_-_26799173 | 4.77 |
ENSDART00000136776
ENSDART00000076484 |
kti12
|
KTI12 chromatin associated homolog |
| chr10_-_34058331 | 4.76 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr6_-_2000017 | 4.74 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr9_+_4382041 | 4.73 |
|
|
|
| KN150623v1_+_258 | 4.71 |
|
|
|
| chr14_-_36004908 | 4.70 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr19_-_27093095 | 4.65 |
|
|
|
| chr12_+_29121368 | 4.63 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr25_+_7067043 | 4.60 |
ENSDART00000179500
|
BX005076.1
|
ENSDARG00000107609 |
| chr16_-_25317921 | 4.60 |
|
|
|
| chr14_-_32937536 | 4.44 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr15_+_29092224 | 4.43 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr20_-_14218080 | 4.42 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr21_-_7334721 | 4.39 |
ENSDART00000136671
|
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr23_-_45107021 | 4.38 |
ENSDART00000076373
|
st8sia7.1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 7.1 |
| chr13_-_21541531 | 4.36 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr13_-_28480530 | 4.36 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr11_+_30005768 | 4.35 |
ENSDART00000167618
|
CR790368.1
|
ENSDARG00000100936 |
| chr14_-_880799 | 4.33 |
ENSDART00000031992
|
rgs14a
|
regulator of G protein signaling 14a |
| chr8_+_41003546 | 4.24 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr16_-_32172641 | 4.22 |
|
|
|
| chr10_-_109097 | 4.21 |
ENSDART00000127228
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr7_+_24257251 | 4.21 |
ENSDART00000136473
|
ENSDARG00000079281
|
ENSDARG00000079281 |
| chr19_+_21209328 | 4.21 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr22_+_25734113 | 4.19 |
ENSDART00000136334
|
si:ch211-250e5.16
|
si:ch211-250e5.16 |
| chr17_+_52526741 | 4.19 |
ENSDART00000109891
|
angel1
|
angel homolog 1 (Drosophila) |
| chr22_-_20377970 | 4.18 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr17_-_30635487 | 4.16 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr22_+_1153361 | 4.15 |
ENSDART00000159761
|
irf6
|
interferon regulatory factor 6 |
| chr7_-_47990610 | 4.14 |
ENSDART00000147968
|
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr16_+_53632289 | 4.12 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| KN150455v1_-_14200 | 4.11 |
|
|
|
| chr15_+_29091983 | 4.11 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr7_+_1337856 | 4.08 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr22_+_22413803 | 4.05 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr14_+_32578103 | 4.03 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr12_-_42212994 | 4.02 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr18_-_40718244 | 4.01 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr4_-_17289857 | 4.00 |
ENSDART00000178686
|
lrmp
|
lymphoid-restricted membrane protein |
| chr20_+_3386573 | 3.98 |
ENSDART00000175369
ENSDART00000176963 ENSDART00000176191 |
CABZ01061478.1
|
ENSDARG00000106218 |
| chr19_+_791656 | 3.92 |
ENSDART00000138406
|
tmem79a
|
transmembrane protein 79a |
| chr3_+_61285878 | 3.91 |
ENSDART00000106266
|
si:zfos-44a5.1
|
si:zfos-44a5.1 |
| chr9_-_28444024 | 3.90 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr15_+_20303539 | 3.89 |
ENSDART00000152473
|
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr5_-_23211957 | 3.85 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr25_+_15901398 | 3.82 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr1_-_44893257 | 3.80 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr20_-_43826667 | 3.77 |
ENSDART00000100637
|
mixl1
|
Mix paired-like homeobox |
| chr14_+_32578253 | 3.76 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr3_-_31747486 | 3.75 |
ENSDART00000076189
|
zgc:171779
|
zgc:171779 |
| chr17_+_25314229 | 3.74 |
ENSDART00000082319
|
tmem54a
|
transmembrane protein 54a |
| chr14_+_31265616 | 3.74 |
|
|
|
| chr7_-_73890600 | 3.73 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr14_-_32937496 | 3.72 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr2_-_9854212 | 3.67 |
ENSDART00000112995
|
wu:fi34b01
|
wu:fi34b01 |
| chr16_-_55211053 | 3.66 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr7_+_39408883 | 3.65 |
ENSDART00000133420
|
tbc1d14
|
TBC1 domain family, member 14 |
| chr19_-_48327666 | 3.63 |
|
|
|
| chr22_+_24112851 | 3.61 |
|
|
|
| chr24_+_34183462 | 3.61 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr12_-_28680035 | 3.57 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr4_-_20456825 | 3.56 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr22_+_25113293 | 3.55 |
ENSDART00000171851
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr7_-_24249672 | 3.53 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr8_-_18637019 | 3.52 |
ENSDART00000172584
ENSDART00000100516 |
stap2b
|
signal transducing adaptor family member 2b |
| chr15_-_34107138 | 3.51 |
ENSDART00000163841
|
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr11_-_13094557 | 3.49 |
ENSDART00000160989
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr12_-_13692190 | 3.47 |
ENSDART00000152370
|
foxh1
|
forkhead box H1 |
| chr15_+_20303330 | 3.47 |
ENSDART00000152473
|
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr16_+_39209567 | 3.41 |
ENSDART00000121756
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr7_+_73447957 | 3.39 |
ENSDART00000050357
ENSDART00000110769 |
btr12
|
bloodthirsty-related gene family, member 12 |
| chr7_-_21571924 | 3.39 |
ENSDART00000166446
|
BX005336.1
|
ENSDARG00000102693 |
| chr22_+_17803309 | 3.39 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr16_-_55210909 | 3.39 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr15_+_34106908 | 3.39 |
|
|
|
| chr18_-_25784873 | 3.38 |
ENSDART00000103046
|
zgc:162879
|
zgc:162879 |
| chr20_-_34894930 | 3.37 |
|
|
|
| KN150699v1_-_15078 | 3.36 |
ENSDART00000159861
|
ENSDARG00000098739
|
ENSDARG00000098739 |
| chr4_+_13902603 | 3.36 |
ENSDART00000137549
|
pphln1
|
periphilin 1 |
| chr25_-_36621626 | 3.35 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr17_-_38939792 | 3.33 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr7_+_39409224 | 3.28 |
ENSDART00000133420
|
tbc1d14
|
TBC1 domain family, member 14 |
| chr15_+_25554119 | 3.26 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| KN150334v1_-_9585 | 3.26 |
ENSDART00000175935
|
CABZ01113810.1
|
ENSDARG00000107898 |
| chr14_-_36004835 | 3.25 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr10_+_43953171 | 3.25 |
|
|
|
| chr16_+_28857531 | 3.22 |
ENSDART00000161525
|
zgc:171704
|
zgc:171704 |
| chr2_+_19514967 | 3.22 |
ENSDART00000166073
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr15_+_44168257 | 3.22 |
ENSDART00000176254
|
CU655961.7
|
ENSDARG00000106724 |
| chr7_+_39409060 | 3.20 |
ENSDART00000133420
|
tbc1d14
|
TBC1 domain family, member 14 |
| chr5_-_14993912 | 3.19 |
ENSDART00000085943
|
taok3a
|
TAO kinase 3a |
| chr7_-_52847458 | 3.18 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr10_-_35205636 | 3.17 |
ENSDART00000131291
|
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr15_-_34106827 | 3.17 |
ENSDART00000163841
|
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr1_+_11508857 | 3.16 |
ENSDART00000172334
ENSDART00000112171 |
tdrd7a
|
tudor domain containing 7 a |
| chr10_+_19625897 | 3.12 |
|
|
|
| chr16_-_25318334 | 3.09 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr21_+_15637516 | 3.09 |
ENSDART00000149371
|
idh3b
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr24_-_31967674 | 3.06 |
ENSDART00000156060
|
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr14_+_25973961 | 3.06 |
ENSDART00000178140
ENSDART00000179471 |
FO082779.2
|
ENSDARG00000106651 |
| chr24_-_10253851 | 3.05 |
ENSDART00000127568
ENSDART00000106260 |
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr10_+_19636552 | 3.05 |
|
|
|
| chr10_-_25448712 | 3.01 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr17_-_23689233 | 2.99 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr15_+_29126271 | 2.99 |
ENSDART00000144546
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr22_+_17803347 | 2.99 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr10_-_1933874 | 2.97 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr10_-_25808033 | 2.97 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| chr7_-_38581532 | 2.96 |
|
|
|
| chr7_-_69014510 | 2.95 |
|
|
|
| chr24_-_30929990 | 2.95 |
|
|
|
| chr22_+_17235696 | 2.95 |
ENSDART00000134798
|
tdrd5
|
tudor domain containing 5 |
| chr16_+_28946580 | 2.94 |
ENSDART00000146525
|
chtopb
|
chromatin target of PRMT1b |
| chr16_+_17807937 | 2.94 |
ENSDART00000149042
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr23_+_43785682 | 2.94 |
ENSDART00000172160
|
ENSDARG00000105093
|
ENSDARG00000105093 |
| chr16_-_42236126 | 2.94 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr1_-_18118467 | 2.92 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr23_+_43868027 | 2.92 |
ENSDART00000112598
ENSDART00000169576 |
otud4
|
OTU deubiquitinase 4 |
| chr7_-_52850565 | 2.92 |
|
|
|
| chr10_+_29247498 | 2.91 |
|
|
|
| chr24_+_34183557 | 2.91 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr23_-_43785497 | 2.91 |
ENSDART00000165963
|
ENSDARG00000102050
|
ENSDARG00000102050 |
| chr18_-_39476656 | 2.90 |
|
|
|
| chr20_-_194135 | 2.89 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr20_-_50089784 | 2.89 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr16_+_53632152 | 2.88 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr20_+_3386522 | 2.88 |
ENSDART00000175369
ENSDART00000176963 ENSDART00000176191 |
CABZ01061478.1
|
ENSDARG00000106218 |
| chr2_+_19515120 | 2.87 |
ENSDART00000166073
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr10_-_36603236 | 2.87 |
ENSDART00000108484
ENSDART00000086861 |
nipblb
|
nipped-B homolog b (Drosophila) |
| chr2_+_32863386 | 2.85 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr20_+_14218237 | 2.84 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr14_-_21320696 | 2.84 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr2_+_26581725 | 2.82 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr1_+_51998406 | 2.82 |
ENSDART00000134658
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr19_-_3547196 | 2.81 |
ENSDART00000166107
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr22_-_25592743 | 2.81 |
ENSDART00000110638
ENSDART00000172092 |
si:ch211-12h2.8
|
si:ch211-12h2.8 |
| chr24_-_20999883 | 2.80 |
ENSDART00000155652
|
gramd1c
|
GRAM domain containing 1c |
| chr7_+_26730804 | 2.79 |
|
|
|
| chr6_+_135255 | 2.78 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr10_-_31336996 | 2.77 |
|
|
|
| chr25_-_8026912 | 2.75 |
|
|
|
| chr7_+_15479700 | 2.75 |
|
|
|
| chr3_-_32699672 | 2.75 |
|
|
|
| chr4_+_16020464 | 2.75 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr25_-_3091345 | 2.74 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr19_+_42661987 | 2.74 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr23_-_45862632 | 2.73 |
ENSDART00000149930
|
cyp17a2
|
cytochrome P450, family 17, subfamily A, polypeptide 2 |
| chr23_-_32267833 | 2.73 |
|
|
|
| chr17_-_25313024 | 2.71 |
ENSDART00000082324
|
zpcx
|
zona pellucida protein C |
| chr18_+_24490621 | 2.70 |
ENSDART00000143108
|
CR847971.1
|
ENSDARG00000091941 |
| chr15_-_28973724 | 2.69 |
|
|
|
| KN150455v1_-_14097 | 2.69 |
|
|
|
| chr7_+_58449127 | 2.68 |
ENSDART00000167166
|
zgc:56231
|
zgc:56231 |
| chr2_-_47766563 | 2.67 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr22_+_15933690 | 2.67 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr13_-_50940551 | 2.67 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 16.8 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 3.1 | 18.7 | GO:0030237 | female sex determination(GO:0030237) |
| 2.7 | 2.7 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 2.6 | 7.7 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 2.1 | 6.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 2.1 | 8.3 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 1.9 | 11.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 1.9 | 5.6 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 1.8 | 12.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 1.8 | 5.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 1.7 | 6.9 | GO:0051151 | regulation of smooth muscle cell differentiation(GO:0051150) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 1.6 | 4.9 | GO:0008347 | glial cell migration(GO:0008347) eurydendroid cell differentiation(GO:0021755) |
| 1.6 | 19.4 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 1.5 | 6.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 1.4 | 4.3 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 1.3 | 7.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 1.1 | 3.4 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 1.1 | 5.5 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 1.1 | 3.2 | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process(GO:1902633) 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process(GO:1902635) |
| 1.0 | 1.0 | GO:0021861 | forebrain radial glial cell differentiation(GO:0021861) |
| 1.0 | 3.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.0 | 3.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 1.0 | 2.9 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 1.0 | 2.9 | GO:0090242 | mesodermal-endodermal cell signaling(GO:0003131) response to vitamin A(GO:0033189) cellular response to vitamin A(GO:0071299) retinoic acid receptor signaling pathway involved in somitogenesis(GO:0090242) |
| 1.0 | 5.7 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.9 | 3.7 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.9 | 24.5 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.9 | 3.6 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.9 | 2.7 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.9 | 2.7 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.8 | 2.5 | GO:0070922 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) small RNA loading onto RISC(GO:0070922) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.8 | 11.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.8 | 3.3 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) negative regulation of skeletal muscle cell proliferation(GO:0014859) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) positive regulation of lamellipodium organization(GO:1902745) |
| 0.8 | 3.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.8 | 3.9 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.8 | 5.4 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.8 | 9.2 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.8 | 3.0 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.7 | 2.9 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.7 | 5.0 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.7 | 2.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.7 | 3.4 | GO:0045719 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.7 | 4.0 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.7 | 4.6 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.7 | 3.9 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.6 | 3.2 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.6 | 0.6 | GO:2000106 | leukocyte apoptotic process(GO:0071887) regulation of leukocyte apoptotic process(GO:2000106) |
| 0.6 | 1.9 | GO:0000493 | box H/ACA snoRNP assembly(GO:0000493) |
| 0.6 | 3.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.6 | 3.2 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.6 | 2.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.6 | 5.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.6 | 0.6 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.6 | 1.8 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.6 | 6.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.6 | 2.3 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.6 | 1.7 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.6 | 0.6 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.6 | 5.7 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.5 | 1.6 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.5 | 3.2 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.5 | 6.3 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.5 | 2.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.5 | 5.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.5 | 2.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.5 | 1.5 | GO:0071295 | cellular response to nutrient(GO:0031670) vitamin D3 metabolic process(GO:0070640) cellular response to vitamin(GO:0071295) cellular response to vitamin D(GO:0071305) |
| 0.5 | 2.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.5 | 1.5 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.5 | 6.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.5 | 1.0 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.5 | 8.2 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.5 | 4.3 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.5 | 2.9 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.5 | 1.4 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) RNA 5'-end processing(GO:0000966) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.5 | 1.4 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.5 | 3.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.5 | 2.3 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.5 | 1.8 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.5 | 2.7 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.4 | 1.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.4 | 1.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.4 | 3.5 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.4 | 1.3 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.4 | 1.3 | GO:0006586 | tryptophan metabolic process(GO:0006568) tryptophan catabolic process(GO:0006569) indolalkylamine metabolic process(GO:0006586) 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) indole-containing compound catabolic process(GO:0042436) anthranilate metabolic process(GO:0043420) indolalkylamine catabolic process(GO:0046218) |
| 0.4 | 5.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.4 | 1.2 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.4 | 6.1 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.4 | 2.4 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.4 | 2.7 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.4 | 1.9 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.4 | 1.5 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.4 | 1.5 | GO:0010662 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.4 | 0.8 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.4 | 12.5 | GO:0048599 | oocyte development(GO:0048599) |
| 0.4 | 3.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.4 | 1.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.4 | 3.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.4 | 2.9 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.4 | 3.9 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.4 | 11.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.4 | 4.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 1.7 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.3 | 2.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.3 | 2.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.3 | 2.7 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 3.7 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.3 | 4.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.3 | 1.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.3 | 2.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 1.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 11.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.3 | 1.0 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.3 | 4.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 1.3 | GO:2000737 | negative regulation of stem cell differentiation(GO:2000737) |
| 0.3 | 1.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.3 | 1.5 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.3 | 1.5 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.3 | 1.2 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.3 | 2.7 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.3 | 2.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.3 | 1.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 8.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.3 | 2.8 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.3 | 5.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.3 | 1.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.3 | 1.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.3 | 2.2 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.3 | 1.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 1.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.3 | 1.1 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.3 | 1.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.3 | 1.1 | GO:0036047 | regulation of ketone biosynthetic process(GO:0010566) protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) |
| 0.3 | 1.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.3 | 2.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.3 | 1.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 1.6 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.3 | 1.8 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.3 | 0.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.3 | 0.8 | GO:0070658 | mechanosensory epithelium regeneration(GO:0070655) mechanoreceptor differentiation involved in mechanosensory epithelium regeneration(GO:0070656) neuromast regeneration(GO:0070657) neuromast hair cell differentiation involved in neuromast regeneration(GO:0070658) |
| 0.3 | 1.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 3.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 4.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.2 | 1.0 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 11.6 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.2 | 1.2 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 2.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 1.4 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 2.4 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.2 | 0.6 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 3.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.2 | 0.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 0.8 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 6.1 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.2 | 0.8 | GO:0045621 | positive regulation of lymphocyte differentiation(GO:0045621) |
| 0.2 | 0.8 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 3.9 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.2 | 2.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 1.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.6 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.2 | 1.8 | GO:0006591 | arginine catabolic process(GO:0006527) ornithine metabolic process(GO:0006591) |
| 0.2 | 5.1 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 5.1 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.2 | 1.6 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.2 | 0.6 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 0.9 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) |
| 0.2 | 0.9 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 1.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 3.5 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 5.1 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.2 | 2.3 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.2 | 4.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.2 | 2.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.2 | 0.5 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.2 | 1.9 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 3.1 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.2 | 0.5 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.2 | 3.7 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.2 | 2.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 1.8 | GO:0046599 | regulation of centriole replication(GO:0046599) metaphase plate congression(GO:0051310) |
| 0.2 | 5.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.2 | 1.0 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.2 | 2.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 4.9 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.2 | 2.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 6.9 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.2 | 0.9 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.1 | 1.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 3.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 1.0 | GO:0000726 | non-recombinational repair(GO:0000726) |
| 0.1 | 3.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.8 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 1.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 2.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.3 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.1 | 0.5 | GO:0021547 | midbrain-hindbrain boundary initiation(GO:0021547) |
| 0.1 | 1.0 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.7 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.1 | 3.0 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 5.7 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 4.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.7 | GO:0035545 | determination of left/right asymmetry in diencephalon(GO:0035462) determination of left/right asymmetry in nervous system(GO:0035545) |
| 0.1 | 2.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.7 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 1.5 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 2.8 | GO:0007492 | endoderm development(GO:0007492) |
| 0.1 | 0.2 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 3.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 3.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.5 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 1.6 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 1.5 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) |
| 0.1 | 2.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 9.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.3 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 1.0 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 0.6 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 0.7 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.6 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.6 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 1.1 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 3.4 | GO:0098661 | inorganic anion transmembrane transport(GO:0098661) |
| 0.1 | 0.6 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 5.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 1.2 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.1 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.1 | 3.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.3 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 1.2 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.5 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling(GO:0098917) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 0.5 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.7 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.7 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.4 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 0.2 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 1.4 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.1 | 1.3 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 0.3 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.7 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.8 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.7 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 6.9 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.1 | 3.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.5 | GO:0044033 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism metabolic process(GO:0044033) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 11.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 3.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 1.4 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 0.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.1 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.1 | 0.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.1 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.1 | 1.1 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 1.1 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 0.2 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.3 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 1.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.5 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.4 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 1.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 1.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.9 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.5 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 0.6 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.4 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.1 | 0.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 2.0 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.1 | 1.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.0 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.1 | 1.7 | GO:1903364 | positive regulation of cellular protein catabolic process(GO:1903364) |
| 0.1 | 1.1 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 | 2.1 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.1 | 1.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 6.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.2 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.5 | GO:0034030 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.5 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.4 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 1.2 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.9 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 2.0 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.7 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 2.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.6 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 2.6 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 12.1 | GO:0006886 | intracellular protein transport(GO:0006886) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 2.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.5 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 1.0 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.0 | 1.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.4 | GO:0032392 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 1.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.3 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 1.3 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 0.5 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0072594 | establishment of protein localization to organelle(GO:0072594) |
| 0.0 | 1.2 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 1.0 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.9 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.2 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0031056 | regulation of histone modification(GO:0031056) |
| 0.0 | 0.1 | GO:0090162 | establishment of apical/basal cell polarity(GO:0035089) establishment of epithelial cell apical/basal polarity(GO:0045198) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0010890 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 0.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.2 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 18.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.1 | 3.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.9 | 2.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.9 | 4.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.8 | 4.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.8 | 2.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.8 | 4.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.8 | 3.2 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.7 | 6.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.7 | 14.1 | GO:0043186 | P granule(GO:0043186) |
| 0.7 | 2.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.7 | 3.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.7 | 4.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.7 | 6.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.7 | 4.6 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.6 | 11.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.6 | 1.8 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.6 | 5.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.6 | 4.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.5 | 3.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.5 | 4.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.5 | 2.6 | GO:0008623 | CHRAC(GO:0008623) |
| 0.5 | 3.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.5 | 5.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.5 | 2.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.5 | 1.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.5 | 2.7 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.5 | 4.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 1.8 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.4 | 1.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 24.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.4 | 5.3 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.4 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.4 | 4.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 1.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 2.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.3 | 3.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.3 | 6.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 8.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.3 | 4.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.3 | 2.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.3 | 14.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.3 | 1.5 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.3 | 5.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 2.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 1.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 0.8 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 1.4 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.2 | 1.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 0.7 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.2 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 7.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 2.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 6.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.2 | 0.4 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.2 | 4.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 8.7 | GO:0030496 | midbody(GO:0030496) |
| 0.2 | 1.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 1.2 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 2.0 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.2 | 9.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.2 | 1.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 1.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 2.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 0.5 | GO:1905202 | methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.2 | 23.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.2 | 1.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.2 | 0.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 2.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.0 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.2 | 11.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.2 | 1.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.0 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 2.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 1.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 10.4 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 6.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.5 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.5 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 2.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 3.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 1.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 2.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 4.0 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 2.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 3.8 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 1.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 3.0 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 0.3 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 3.8 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 2.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.5 | GO:0032155 | cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0030670 | endocytic vesicle membrane(GO:0030666) phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 24.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 2.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 8.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 3.0 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.0 | 0.2 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.4 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.2 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.0 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.9 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 10.8 | GO:0004508 | steroid 17-alpha-monooxygenase activity(GO:0004508) 17-alpha-hydroxyprogesterone aldolase activity(GO:0047442) |
| 2.1 | 6.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.8 | 10.6 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 1.2 | 4.9 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 1.2 | 4.8 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 1.1 | 5.6 | GO:0070697 | activin receptor binding(GO:0070697) |
| 1.1 | 3.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.1 | 3.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 1.1 | 6.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 1.1 | 4.2 | GO:0072572 | poly-ADP-D-ribose binding(GO:0072572) |
| 1.0 | 3.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 1.0 | 3.1 | GO:0052907 | U6 snRNA 3'-end binding(GO:0030629) 23S rRNA (adenine(1618)-N(6))-methyltransferase activity(GO:0052907) |
| 1.0 | 2.9 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.0 | 3.9 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 1.0 | 5.7 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.9 | 2.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.9 | 3.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.9 | 3.7 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.9 | 24.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.9 | 8.2 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.9 | 3.6 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.9 | 2.7 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.9 | 2.6 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.8 | 3.4 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.8 | 8.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.8 | 4.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.8 | 2.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.8 | 2.4 | GO:0016843 | amine-lyase activity(GO:0016843) strictosidine synthase activity(GO:0016844) |
| 0.7 | 7.9 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.7 | 2.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.7 | 7.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.7 | 2.7 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.6 | 1.9 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.6 | 3.1 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.6 | 3.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.6 | 2.5 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.6 | 3.6 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.6 | 3.5 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.6 | 1.8 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.6 | 1.7 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.6 | 2.9 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.6 | 2.9 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.6 | 2.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.6 | 6.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.6 | 1.7 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.6 | 1.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 1.6 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.5 | 5.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.5 | 10.0 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.5 | 6.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.5 | 3.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.5 | 5.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.5 | 9.3 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.5 | 1.5 | GO:0016595 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.5 | 13.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.5 | 1.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.5 | 1.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.5 | 2.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.4 | 6.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 1.3 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.4 | 2.2 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.4 | 5.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.4 | 1.3 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.4 | 23.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.4 | 1.6 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.4 | 3.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.4 | 3.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.4 | 1.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.4 | 1.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.4 | 3.0 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.4 | 2.9 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.4 | 2.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.4 | 1.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.4 | 1.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 1.0 | GO:0016422 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.3 | 3.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 4.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 1.0 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.3 | 6.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 1.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.3 | 1.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.3 | 1.0 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.3 | 5.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.3 | 1.2 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.3 | 0.9 | GO:0043531 | ADP binding(GO:0043531) |
| 0.3 | 4.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.3 | 1.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.3 | 1.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 1.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 0.9 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.3 | 8.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 5.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.3 | 1.7 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.3 | 0.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 1.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 0.8 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.3 | 5.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 2.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 5.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.3 | 1.1 | GO:0061697 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.3 | 8.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.3 | 1.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 2.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.2 | 0.7 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.2 | 2.2 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.2 | 3.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 0.9 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 1.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 1.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 1.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 1.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 3.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 3.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.6 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.2 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 0.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.6 | GO:0048270 | methionine adenosyltransferase regulator activity(GO:0048270) |
| 0.2 | 1.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 1.0 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.2 | 3.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.0 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.2 | 1.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 0.6 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 2.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 12.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 3.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 1.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 0.5 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 1.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 0.5 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.2 | 3.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 1.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 2.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 6.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 0.8 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 11.3 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.2 | 2.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 1.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.6 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 1.9 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 5.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.6 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.5 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 2.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 1.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.4 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 2.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.0 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 1.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 2.5 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.5 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 27.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.6 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 1.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 2.4 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 1.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 1.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.1 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.3 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.1 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.4 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.3 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 1.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.3 | GO:0005460 | UDP-glucose transmembrane transporter activity(GO:0005460) |
| 0.1 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.1 | 1.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 1.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.5 | GO:0015245 | long-chain fatty acid transporter activity(GO:0005324) fatty acid transporter activity(GO:0015245) |
| 0.1 | 12.1 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 0.8 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.2 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 4.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 8.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 2.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 17.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 4.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 2.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 3.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 1.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 1.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 3.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.6 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 1.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 6.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.5 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 14.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.5 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.4 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.5 | GO:0015296 | anion:cation symporter activity(GO:0015296) |
| 0.0 | 3.5 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 0.8 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 2.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.1 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.3 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.3 | GO:0051287 | NAD binding(GO:0051287) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 12.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.4 | 9.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.4 | 2.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 5.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 2.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 2.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 6.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 8.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 1.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 3.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 1.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 3.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 6.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 6.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 0.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 0.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.8 | 2.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.7 | 3.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.6 | 10.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.6 | 3.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.6 | 11.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.5 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.5 | 3.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.5 | 7.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.4 | 1.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.4 | 3.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 1.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.4 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 6.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.4 | 4.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 3.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.3 | 5.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 1.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 1.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.3 | 1.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 7.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 2.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 6.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 1.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 4.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 1.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 2.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 3.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 0.7 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.2 | 2.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 2.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 4.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 3.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 6.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 4.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.4 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 1.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 5.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 3.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 1.8 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 1.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 10.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 5.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.8 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 0.6 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 3.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.0 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |