Project
DANIO-CODE
Navigation
Downloads
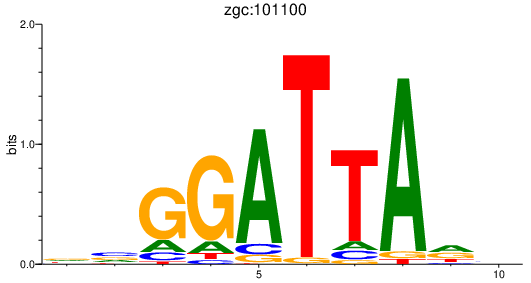
Results for zgc:101100
Z-value: 0.52
Transcription factors associated with zgc:101100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zgc
|
ENSDARG00000021959 | 101100 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zgc:101100 | dr10_dc_chr8_+_20456215_20456242 | -0.57 | 2.0e-02 | Click! |
Activity profile of zgc:101100 motif
Sorted Z-values of zgc:101100 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zgc:101100
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_20670164 | 1.18 |
ENSDART00000169077
|
org
|
oogenesis-related gene |
| chr25_-_20932941 | 1.00 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr4_-_3184182 | 0.95 |
|
|
|
| chr19_-_42668754 | 0.78 |
ENSDART00000132591
|
si:ch211-191i18.4
|
si:ch211-191i18.4 |
| chr5_-_23525132 | 0.70 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr2_+_50680126 | 0.69 |
ENSDART00000122716
|
ENSDARG00000090398
|
ENSDARG00000090398 |
| chr2_+_30800532 | 0.60 |
|
|
|
| chr19_+_48254284 | 0.51 |
|
|
|
| chr16_+_53632289 | 0.49 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr3_+_34690515 | 0.49 |
|
|
|
| chr13_-_50299643 | 0.45 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr1_-_44227583 | 0.44 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr3_+_34690574 | 0.44 |
|
|
|
| chr3_+_18657831 | 0.43 |
ENSDART00000055757
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr5_-_23525331 | 0.41 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr2_+_55112111 | 0.41 |
ENSDART00000163834
|
rnf126
|
ring finger protein 126 |
| chr1_-_54391298 | 0.41 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr4_+_30530 | 0.41 |
ENSDART00000157825
|
syn3
|
synapsin III |
| chr19_+_24483983 | 0.39 |
ENSDART00000141351
|
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr19_+_48254224 | 0.38 |
|
|
|
| chr14_+_30413625 | 0.38 |
ENSDART00000172778
|
yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr10_+_8670564 | 0.36 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr13_+_24416967 | 0.36 |
ENSDART00000177677
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr18_-_43890836 | 0.36 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr3_+_33608669 | 0.35 |
|
|
|
| chr7_+_66660882 | 0.35 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr23_-_19055201 | 0.34 |
ENSDART00000147617
|
bcl2l1
|
bcl2-like 1 |
| chr2_-_9898351 | 0.32 |
ENSDART00000165712
|
selt1a
|
selenoprotein T, 1a |
| chr10_+_2813945 | 0.32 |
ENSDART00000131505
|
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr7_-_18364873 | 0.32 |
ENSDART00000172419
|
si:ch211-119e14.1
|
si:ch211-119e14.1 |
| chr13_-_50299821 | 0.31 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr3_+_33608622 | 0.31 |
|
|
|
| chr6_-_13215216 | 0.31 |
ENSDART00000112883
|
fmnl2b
|
formin-like 2b |
| chr10_+_2813985 | 0.31 |
ENSDART00000055869
|
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr16_+_53632152 | 0.30 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr16_+_29651573 | 0.30 |
ENSDART00000149520
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr2_-_9898419 | 0.30 |
ENSDART00000081214
ENSDART00000171529 |
selt1a
|
selenoprotein T, 1a |
| chr8_-_49227143 | 0.30 |
|
|
|
| chr24_+_9740978 | 0.29 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr11_+_30049041 | 0.28 |
|
|
|
| chr11_-_2437361 | 0.28 |
|
|
|
| chr16_+_25096821 | 0.26 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr16_+_53632229 | 0.26 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr24_-_20782338 | 0.26 |
ENSDART00000130958
|
ccdc58
|
coiled-coil domain containing 58 |
| chr11_-_2437396 | 0.26 |
|
|
|
| chr6_-_13215294 | 0.25 |
ENSDART00000112883
|
fmnl2b
|
formin-like 2b |
| chr16_+_11406779 | 0.25 |
ENSDART00000125158
|
znf574
|
zinc finger protein 574 |
| chr14_-_47408279 | 0.24 |
ENSDART00000083833
|
bbs7
|
Bardet-Biedl syndrome 7 |
| chr24_+_16861210 | 0.24 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr11_+_31417477 | 0.24 |
ENSDART00000139900
|
diaph3
|
diaphanous-related formin 3 |
| chr20_+_35156812 | 0.24 |
|
|
|
| chr25_+_181958 | 0.23 |
ENSDART00000155412
|
RPS17
|
ribosomal protein S17 |
| chr4_+_43810061 | 0.23 |
ENSDART00000157214
|
si:dkey-7j22.2
|
si:dkey-7j22.2 |
| chr19_+_24484213 | 0.23 |
ENSDART00000100420
|
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr11_+_30049139 | 0.23 |
|
|
|
| chr11_+_16439987 | 0.22 |
ENSDART00000104072
|
kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr1_-_44227620 | 0.21 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr12_+_33359784 | 0.20 |
ENSDART00000007053
|
narf
|
nuclear prelamin A recognition factor |
| chr15_-_26954009 | 0.20 |
ENSDART00000156292
|
si:dkey-243i1.1
|
si:dkey-243i1.1 |
| chr20_+_34768464 | 0.19 |
ENSDART00000152836
|
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr14_+_21854812 | 0.18 |
ENSDART00000082697
ENSDART00000054982 |
gabra6a
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6a |
| chr19_-_42668644 | 0.17 |
ENSDART00000151022
|
si:ch211-191i18.4
|
si:ch211-191i18.4 |
| chr13_+_38688704 | 0.15 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr6_-_43273456 | 0.14 |
|
|
|
| chr8_-_10978942 | 0.14 |
ENSDART00000064066
|
bcas2
|
BCAS2, pre-mRNA processing factor |
| chr18_-_38263768 | 0.14 |
ENSDART00000076399
|
nat10
|
N-acetyltransferase 10 |
| chr20_-_18836698 | 0.14 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr11_+_30972891 | 0.14 |
ENSDART00000129393
|
trmt1
|
tRNA methyltransferase 1 |
| chr3_+_33608743 | 0.13 |
|
|
|
| chr14_+_5554581 | 0.13 |
ENSDART00000041279
|
tubb4b
|
tubulin, beta 4B class IVb |
| chr5_+_69369417 | 0.13 |
|
|
|
| chr21_-_9469489 | 0.12 |
ENSDART00000162225
ENSDART00000163874 ENSDART00000168173 |
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr14_-_9274939 | 0.12 |
|
|
|
| chr3_-_23382550 | 0.11 |
ENSDART00000170200
|
BX682558.1
|
ENSDARG00000101975 |
| chr14_+_30413350 | 0.10 |
ENSDART00000009385
|
yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr6_-_33940201 | 0.10 |
ENSDART00000132762
|
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr18_+_17765358 | 0.10 |
ENSDART00000137239
|
si:ch211-216l23.2
|
si:ch211-216l23.2 |
| chr22_+_34726990 | 0.09 |
ENSDART00000082066
|
atpv0e2
|
ATPase, H+ transporting V0 subunit e2 |
| chr22_+_35299551 | 0.08 |
ENSDART00000165353
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr8_+_2597651 | 0.08 |
ENSDART00000165943
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr15_+_43038899 | 0.08 |
ENSDART00000152222
|
acsl3a
|
acyl-CoA synthetase long-chain family member 3a |
| chr5_-_28025557 | 0.07 |
ENSDART00000131729
|
tnc
|
tenascin C |
| chr3_+_34690823 | 0.06 |
|
|
|
| chr5_+_25599519 | 0.06 |
ENSDART00000144303
|
ENSDARG00000035573
|
ENSDARG00000035573 |
| chr17_-_49910160 | 0.05 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr23_-_77607 | 0.04 |
|
|
|
| chr17_-_15141309 | 0.04 |
ENSDART00000103405
|
gch1
|
GTP cyclohydrolase 1 |
| chr5_+_58824344 | 0.04 |
ENSDART00000148727
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr6_+_30385605 | 0.03 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr12_-_10667591 | 0.03 |
ENSDART00000164038
|
ENSDARG00000101171
|
ENSDARG00000101171 |
| chr19_-_41318931 | 0.03 |
ENSDART00000004514
|
bet1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr24_-_24859135 | 0.03 |
ENSDART00000136860
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr4_-_11054645 | 0.02 |
ENSDART00000067262
|
mettl25
|
methyltransferase like 25 |
| chr15_+_2978541 | 0.02 |
ENSDART00000110613
|
lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr16_+_25096883 | 0.01 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr7_-_17527827 | 0.01 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 0.9 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 1.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.7 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.4 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.4 | GO:0051281 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.1 | 0.8 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.6 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.1 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.2 | GO:0060078 | synaptic transmission, GABAergic(GO:0051932) regulation of postsynaptic membrane potential(GO:0060078) |
| 0.0 | 0.1 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.0 | GO:0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0071007 | Prp19 complex(GO:0000974) U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.4 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 1.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |