Project
DANIO-CODE
Navigation
Downloads
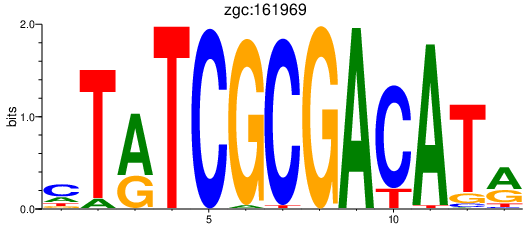
Results for zgc:161969
Z-value: 0.68
Transcription factors associated with zgc:161969
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zgc
|
ENSDARG00000041359 | 161969 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zgc:161969 | dr10_dc_chr6_+_16341787_16341900 | -0.73 | 1.4e-03 | Click! |
Activity profile of zgc:161969 motif
Sorted Z-values of zgc:161969 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zgc:161969
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_53850871 | 1.66 |
ENSDART00000158149
|
tmem203
|
transmembrane protein 203 |
| chr11_-_44107913 | 1.47 |
|
|
|
| chr8_-_19236074 | 1.34 |
ENSDART00000137994
|
zgc:77486
|
zgc:77486 |
| chr5_-_26164683 | 1.13 |
ENSDART00000140392
|
rnf181
|
ring finger protein 181 |
| chr8_-_19235896 | 1.10 |
ENSDART00000036148
|
zgc:77486
|
zgc:77486 |
| chr5_-_23741624 | 0.97 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr5_-_26164728 | 0.91 |
ENSDART00000140392
|
rnf181
|
ring finger protein 181 |
| chr5_-_23741680 | 0.80 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr9_-_34700736 | 0.77 |
ENSDART00000169114
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr9_+_3576854 | 0.77 |
ENSDART00000147650
|
tlk1a
|
tousled-like kinase 1a |
| chr21_+_26486084 | 0.75 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr21_+_19958025 | 0.73 |
|
|
|
| chr15_-_26153718 | 0.71 |
ENSDART00000140214
|
dph1
|
diphthamide biosynthesis 1 |
| chr8_-_19235793 | 0.63 |
ENSDART00000036148
|
zgc:77486
|
zgc:77486 |
| chr5_-_23741334 | 0.61 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr20_+_26005068 | 0.58 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr16_-_26201059 | 0.54 |
ENSDART00000148653
ENSDART00000148923 |
tmem145
|
transmembrane protein 145 |
| chr14_-_33084418 | 0.49 |
ENSDART00000105688
|
nkap
|
NFKB activating protein |
| chr21_+_26485851 | 0.47 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr8_+_52297522 | 0.41 |
ENSDART00000098439
|
ube2d4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr5_+_26165043 | 0.29 |
ENSDART00000144169
|
si:ch211-102c2.8
|
si:ch211-102c2.8 |
| chr14_-_33084808 | 0.26 |
ENSDART00000001318
|
nkap
|
NFKB activating protein |
| chr21_+_26486469 | 0.25 |
ENSDART00000145467
|
adssl
|
adenylosuccinate synthase, like |
| chr22_-_29690871 | 0.16 |
ENSDART00000059882
|
shoc2
|
SHOC2 leucine-rich repeat scaffold protein |
| chr3_+_34104493 | 0.00 |
ENSDART00000124631
|
znf207a
|
zinc finger protein 207, a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0030730 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.4 | 2.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.3 | 1.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.7 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.6 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |