Project
DANIO-CODE
Navigation
Downloads
Results for zic1_si:dkey-149i17.7
Z-value: 1.13
Transcription factors associated with zic1_si:dkey-149i17.7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
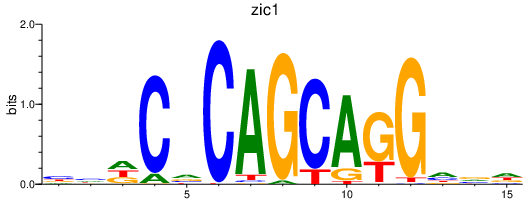
zic1
|
ENSDARG00000015567 | zic family member 1 (odd-paired homolog, Drosophila) |
|
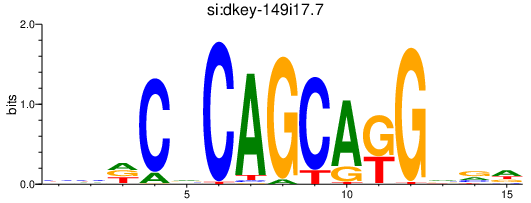
si_dkey-149i17.7
|
ENSDARG00000094684 | si_dkey-149i17.7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zic1 | dr10_dc_chr24_-_4941954_4941994 | 0.35 | 1.8e-01 | Click! |
Activity profile of zic1_si:dkey-149i17.7 motif
Sorted Z-values of zic1_si:dkey-149i17.7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zic1_si:dkey-149i17.7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_39130411 | 2.25 |
ENSDART00000144750
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr13_-_44706874 | 2.03 |
ENSDART00000140442
|
si:dkeyp-2e4.3
|
si:dkeyp-2e4.3 |
| chr5_-_7666964 | 2.03 |
ENSDART00000167643
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr22_+_17091411 | 2.01 |
ENSDART00000170076
|
frem1b
|
Fras1 related extracellular matrix 1b |
| chr5_-_54954988 | 1.88 |
ENSDART00000142912
|
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr1_+_44205158 | 1.83 |
ENSDART00000142820
|
wu:fc21g02
|
wu:fc21g02 |
| chr23_-_46250905 | 1.63 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr19_-_27807312 | 1.61 |
|
|
|
| KN149946v1_-_12861 | 1.56 |
ENSDART00000163016
|
CABZ01048402.2
|
ENSDARG00000103875 |
| chr12_+_14176424 | 1.40 |
ENSDART00000078052
|
mto1
|
mitochondrial tRNA translation optimization 1 |
| chr13_-_50299643 | 1.38 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr14_-_223462 | 1.27 |
|
|
|
| chr23_-_46251021 | 1.24 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr2_+_25363717 | 1.24 |
ENSDART00000142601
|
stag1a
|
stromal antigen 1a |
| chr22_+_4722336 | 1.13 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr3_-_39313029 | 1.10 |
ENSDART00000156123
|
CU464118.1
|
ENSDARG00000097747 |
| chr4_-_26333625 | 1.09 |
ENSDART00000170709
|
st8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr11_+_30049795 | 1.08 |
|
|
|
| chr15_-_4500676 | 1.07 |
|
|
|
| chr3_-_32727588 | 1.07 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr19_+_1356824 | 1.04 |
ENSDART00000081779
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
| chr13_+_30781652 | 0.98 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr1_-_343097 | 0.97 |
ENSDART00000010092
|
gas6
|
growth arrest-specific 6 |
| chr9_+_308260 | 0.97 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr18_+_34205362 | 0.94 |
ENSDART00000130831
|
gmps
|
guanine monophosphate synthase |
| chr20_+_26993709 | 0.94 |
ENSDART00000133293
|
ftr97
|
finTRIM family, member 97 |
| chr8_-_43151611 | 0.94 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr14_-_45887823 | 0.93 |
ENSDART00000178932
ENSDART00000160657 |
cltb
|
clathrin, light chain B |
| chr5_+_69096325 | 0.92 |
ENSDART00000014649
|
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr19_-_24859287 | 0.89 |
ENSDART00000163763
|
thbs3b
|
thrombospondin 3b |
| chr14_-_2780569 | 0.88 |
ENSDART00000090213
|
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr17_+_24428093 | 0.86 |
ENSDART00000131417
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr14_+_51850006 | 0.86 |
|
|
|
| chr8_+_24276667 | 0.84 |
|
|
|
| chr23_+_32573474 | 0.83 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr9_-_56692366 | 0.82 |
|
|
|
| chr14_-_2780615 | 0.81 |
ENSDART00000090213
|
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr3_+_28450576 | 0.81 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr25_-_352985 | 0.80 |
ENSDART00000165705
|
szl
|
sizzled |
| chr8_+_21974863 | 0.78 |
|
|
|
| chr8_+_24276581 | 0.78 |
|
|
|
| chr14_+_51850088 | 0.76 |
|
|
|
| chr15_-_10345337 | 0.74 |
|
|
|
| chr5_+_52198613 | 0.74 |
ENSDART00000162459
|
scarb2a
|
scavenger receptor class B, member 2a |
| chr5_-_71394207 | 0.71 |
ENSDART00000050971
|
rab14l
|
RAB14, member RAS oncogene family, like |
| chr2_-_58214171 | 0.71 |
ENSDART00000169909
ENSDART00000168590 |
ENSDARG00000103284
|
ENSDARG00000103284 |
| chr19_-_2314582 | 0.70 |
ENSDART00000166669
|
btd
|
biotinidase |
| chr10_-_20524586 | 0.70 |
|
|
|
| chr11_-_9479689 | 0.69 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr16_-_55320569 | 0.68 |
ENSDART00000156368
|
ENSDARG00000069583
|
ENSDARG00000069583 |
| chr7_-_24428781 | 0.67 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr11_-_22966274 | 0.67 |
ENSDART00000003646
|
optc
|
opticin |
| chr8_-_24276524 | 0.65 |
|
|
|
| chr23_-_46251093 | 0.64 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr12_-_48643687 | 0.64 |
ENSDART00000178135
ENSDART00000176040 |
CABZ01078412.1
|
ENSDARG00000107260 |
| chr13_+_30781718 | 0.64 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr25_+_34340352 | 0.64 |
ENSDART00000163190
|
tmem231
|
transmembrane protein 231 |
| chr5_-_20417926 | 0.63 |
ENSDART00000158030
|
si:ch211-225b11.4
|
si:ch211-225b11.4 |
| chr24_-_34794463 | 0.63 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr7_-_18629056 | 0.62 |
ENSDART00000021502
|
mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr14_+_51850213 | 0.62 |
|
|
|
| chr14_+_51848088 | 0.61 |
ENSDART00000168874
|
rpl26
|
ribosomal protein L26 |
| chr12_-_13690539 | 0.61 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr13_-_25502297 | 0.61 |
ENSDART00000077627
ENSDART00000139237 |
ret
|
ret proto-oncogene receptor tyrosine kinase |
| chr11_+_30049412 | 0.60 |
|
|
|
| chr5_-_21520111 | 0.60 |
|
|
|
| chr11_-_9479592 | 0.59 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr25_-_28691412 | 0.58 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr8_-_12171624 | 0.58 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr12_+_48643983 | 0.58 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr23_-_4175790 | 0.56 |
ENSDART00000109807
|
ENSDARG00000076299
|
ENSDARG00000076299 |
| chr18_-_45892 | 0.54 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr15_-_27778111 | 0.54 |
ENSDART00000134373
|
lhx1a
|
LIM homeobox 1a |
| chr7_+_22272746 | 0.54 |
ENSDART00000101528
|
chrnb1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr25_+_36864779 | 0.53 |
ENSDART00000171602
|
chmp1a
|
charged multivesicular body protein 1A |
| chr19_-_14070901 | 0.53 |
ENSDART00000158059
|
trnau1apa
|
tRNA selenocysteine 1 associated protein 1a |
| chr6_+_10141622 | 0.52 |
ENSDART00000175899
|
BX005368.2
|
ENSDARG00000106412 |
| chr17_+_1799209 | 0.51 |
ENSDART00000112183
|
cep170b
|
centrosomal protein 170B |
| chr25_-_35530474 | 0.50 |
|
|
|
| chr4_+_12932950 | 0.50 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr4_+_12932838 | 0.49 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr22_+_12406830 | 0.49 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr15_-_1225595 | 0.49 |
|
|
|
| chr2_+_4562038 | 0.48 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr20_+_9776103 | 0.48 |
ENSDART00000053834
|
psmc6
|
proteasome 26S subunit, ATPase 6 |
| KN150487v1_+_15409 | 0.48 |
ENSDART00000166996
|
CABZ01074304.1
|
ENSDARG00000100224 |
| chr23_+_9285446 | 0.47 |
ENSDART00000033663
|
rps21
|
ribosomal protein S21 |
| chr12_-_8780098 | 0.47 |
ENSDART00000113148
|
jmjd1cb
|
jumonji domain containing 1Cb |
| chr18_+_22285979 | 0.46 |
|
|
|
| chr25_-_10930008 | 0.46 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr4_-_16461748 | 0.46 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr10_-_2498928 | 0.45 |
ENSDART00000081926
|
CU856539.1
|
ENSDARG00000010023 |
| chr2_-_37151666 | 0.45 |
ENSDART00000146123
|
elavl1
|
ELAV like RNA binding protein 1 |
| chr25_-_36864478 | 0.44 |
ENSDART00000157623
|
pdhx
|
pyruvate dehydrogenase complex, component X |
| chr9_-_137883 | 0.44 |
ENSDART00000169377
|
FQ377903.3
|
ENSDARG00000104933 |
| chr13_-_51662834 | 0.43 |
|
|
|
| chr4_-_16461881 | 0.43 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr14_+_6834563 | 0.42 |
ENSDART00000170994
ENSDART00000129898 |
ctsf
|
cathepsin F |
| chr13_-_48465147 | 0.42 |
ENSDART00000132895
|
ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr18_+_5456221 | 0.42 |
|
|
|
| chr9_-_17278574 | 0.42 |
ENSDART00000039157
|
mycbp2
|
MYC binding protein 2 |
| chr6_-_29753229 | 0.42 |
|
|
|
| chr18_+_2203805 | 0.42 |
|
|
|
| chr24_+_42884 | 0.41 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr7_-_40385050 | 0.40 |
ENSDART00000142315
|
nom1
|
nucleolar protein with MIF4G domain 1 |
| chr13_+_1227220 | 0.40 |
ENSDART00000114907
|
ifngr1
|
interferon gamma receptor 1 |
| chr18_-_47026460 | 0.40 |
ENSDART00000108574
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr13_-_36996246 | 0.39 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr25_-_35698109 | 0.39 |
ENSDART00000134928
|
nfatc3b
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3b |
| chr10_-_2499073 | 0.39 |
ENSDART00000081926
|
CU856539.1
|
ENSDARG00000010023 |
| chr9_-_6230087 | 0.38 |
ENSDART00000032368
|
tmtops2b
|
teleost multiple tissue opsin 2b |
| chr20_-_11179880 | 0.38 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr11_-_176933 | 0.38 |
ENSDART00000172920
|
cdk4
|
cyclin-dependent kinase 4 |
| chr10_+_22948846 | 0.38 |
ENSDART00000167874
|
zgc:103508
|
zgc:103508 |
| chr14_-_45887648 | 0.38 |
ENSDART00000074208
|
cltb
|
clathrin, light chain B |
| chr7_-_18629119 | 0.37 |
ENSDART00000021502
|
mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr11_+_268997 | 0.37 |
ENSDART00000168188
|
mettl1
|
methyltransferase like 1 |
| chr17_-_15659244 | 0.36 |
ENSDART00000142972
|
manea
|
mannosidase, endo-alpha |
| chr15_-_30975632 | 0.36 |
|
|
|
| chr24_-_39884167 | 0.36 |
ENSDART00000087441
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr25_-_16881637 | 0.36 |
ENSDART00000133897
|
ccnd2a
|
cyclin D2, a |
| chr5_-_54955163 | 0.36 |
ENSDART00000133156
|
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr20_+_34126950 | 0.36 |
ENSDART00000134305
|
prg4b
|
proteoglycan 4b |
| chr12_-_9046799 | 0.36 |
ENSDART00000125230
|
exoc6
|
exocyst complex component 6 |
| chr1_+_39844690 | 0.36 |
ENSDART00000122059
|
scoca
|
short coiled-coil protein a |
| chr3_-_40020899 | 0.35 |
ENSDART00000025285
|
drg2
|
developmentally regulated GTP binding protein 2 |
| chr22_+_12406922 | 0.35 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr2_+_34198009 | 0.35 |
|
|
|
| chr17_+_45932080 | 0.35 |
ENSDART00000157326
|
kif26ab
|
kinesin family member 26Ab |
| chr6_-_907747 | 0.35 |
ENSDART00000171313
|
orc4
|
origin recognition complex, subunit 4 |
| chr19_-_47366517 | 0.35 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr20_+_52778914 | 0.35 |
ENSDART00000138641
|
si:dkey-235d18.5
|
si:dkey-235d18.5 |
| chr7_+_36267647 | 0.35 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr3_+_36497377 | 0.34 |
|
|
|
| chr9_-_2938937 | 0.34 |
|
|
|
| chr1_+_29926606 | 0.34 |
|
|
|
| chr21_-_13133786 | 0.33 |
ENSDART00000023834
|
setb
|
SET nuclear proto-oncogene b |
| chr3_-_18426055 | 0.32 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr9_-_56692281 | 0.32 |
|
|
|
| chr4_-_18862349 | 0.32 |
ENSDART00000021782
|
mcat
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr8_-_45422915 | 0.31 |
ENSDART00000150067
|
ywhabb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide b |
| chr20_-_11179958 | 0.31 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr6_-_16156494 | 0.31 |
|
|
|
| chr25_-_31305819 | 0.31 |
|
|
|
| chr17_+_24045786 | 0.31 |
ENSDART00000132755
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr23_+_27985224 | 0.31 |
ENSDART00000171859
|
ENSDARG00000100606
|
ENSDARG00000100606 |
| chr16_-_49672970 | 0.30 |
ENSDART00000127513
|
znf385d
|
zinc finger protein 385D |
| chr7_-_13638486 | 0.30 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr6_-_51538315 | 0.30 |
|
|
|
| chr19_-_32487376 | 0.30 |
|
|
|
| chr2_-_48103519 | 0.30 |
ENSDART00000056305
|
fzd8b
|
frizzled class receptor 8b |
| chr7_+_16696469 | 0.29 |
|
|
|
| chr23_+_28792316 | 0.29 |
ENSDART00000078156
|
srm
|
spermidine synthase |
| chr16_-_36022327 | 0.29 |
ENSDART00000172252
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr11_-_269181 | 0.29 |
|
|
|
| chr19_+_38033219 | 0.28 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr14_-_25645760 | 0.28 |
ENSDART00000141916
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr15_+_20303330 | 0.28 |
ENSDART00000152473
|
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr24_-_16772614 | 0.27 |
ENSDART00000110715
|
cmbl
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr1_-_40587797 | 0.27 |
ENSDART00000170895
|
rgs12b
|
regulator of G protein signaling 12b |
| chr14_+_23220869 | 0.26 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr7_-_24723345 | 0.26 |
ENSDART00000002961
|
rcor2
|
REST corepressor 2 |
| chr1_-_48900185 | 0.24 |
ENSDART00000113072
|
zgc:163136
|
zgc:163136 |
| chr19_-_10996809 | 0.24 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr23_-_27621359 | 0.24 |
ENSDART00000144419
|
larp4aa
|
La ribonucleoprotein domain family, member 4Aa |
| KN150487v1_+_15682 | 0.23 |
ENSDART00000166996
|
CABZ01074304.1
|
ENSDARG00000100224 |
| chr17_+_24427354 | 0.23 |
ENSDART00000163221
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr17_-_49355906 | 0.23 |
ENSDART00000162563
|
znf292a
|
zinc finger protein 292a |
| chr23_+_32465915 | 0.23 |
ENSDART00000171715
ENSDART00000157514 ENSDART00000175644 ENSDART00000179137 |
taf8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr15_+_20303673 | 0.23 |
ENSDART00000152473
|
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr14_-_17282615 | 0.23 |
ENSDART00000006716
ENSDART00000136242 |
selt2
|
selenoprotein T, 2 |
| chr14_+_27936303 | 0.23 |
ENSDART00000003293
|
mid2
|
midline 2 |
| chr17_+_24045582 | 0.23 |
ENSDART00000132755
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr2_-_47827229 | 0.23 |
ENSDART00000056882
|
cul3a
|
cullin 3a |
| chr25_-_20994084 | 0.23 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| chr5_+_68055581 | 0.22 |
ENSDART00000061406
|
usp39
|
ubiquitin specific peptidase 39 |
| chr1_-_58606982 | 0.22 |
ENSDART00000158067
|
txndc11
|
thioredoxin domain containing 11 |
| chr7_+_10289579 | 0.22 |
|
|
|
| chr6_+_17928720 | 0.22 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr1_-_55929842 | 0.21 |
ENSDART00000133693
|
CABZ01059412.1
|
ENSDARG00000093082 |
| chr20_-_53040549 | 0.21 |
ENSDART00000171177
ENSDART00000040265 |
tbc1d7
|
TBC1 domain family, member 7 |
| chr22_-_20963723 | 0.20 |
|
|
|
| chr17_-_27030048 | 0.20 |
ENSDART00000050018
|
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr17_+_24803152 | 0.20 |
ENSDART00000112313
|
wdr43
|
WD repeat domain 43 |
| chr8_+_6989445 | 0.20 |
ENSDART00000134440
|
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr13_+_12651778 | 0.19 |
ENSDART00000090000
|
si:ch211-233a24.2
|
si:ch211-233a24.2 |
| chr21_+_45629613 | 0.19 |
|
|
|
| chr23_+_12799718 | 0.18 |
|
|
|
| chr15_+_1018191 | 0.18 |
|
|
|
| chr18_-_45750 | 0.18 |
ENSDART00000148821
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr22_+_39012456 | 0.18 |
ENSDART00000176985
|
CABZ01044615.1
|
ENSDARG00000108810 |
| chr2_+_7341600 | 0.18 |
ENSDART00000146547
|
ENSDARG00000078299
|
ENSDARG00000078299 |
| chr25_-_20993989 | 0.17 |
|
|
|
| chr16_+_42067930 | 0.17 |
ENSDART00000102789
|
fli1b
|
Fli-1 proto-oncogene, ETS transcription factor b |
| chr23_+_17940659 | 0.17 |
|
|
|
| chr10_-_36864268 | 0.17 |
ENSDART00000165853
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr8_-_11732343 | 0.16 |
ENSDART00000091684
|
anapc7
|
anaphase promoting complex subunit 7 |
| chr7_-_20556921 | 0.16 |
ENSDART00000026098
|
dnajc3b
|
DnaJ (Hsp40) homolog, subfamily C, member 3b |
| chr23_-_28766019 | 0.16 |
ENSDART00000078141
|
ribc1
|
RIB43A domain with coiled-coils 1 |
| chr22_+_2681009 | 0.16 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.3 | 3.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 1.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 1.0 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.2 | 0.7 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.2 | 0.7 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.2 | 1.1 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 0.5 | GO:0097378 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.1 | 0.7 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 1.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 1.4 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.6 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.5 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.4 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.6 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 2.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.9 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 1.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.4 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.5 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.2 | GO:0048855 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.3 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 1.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.5 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 1.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.6 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.2 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.4 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.8 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 1.0 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.5 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 2.0 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.3 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.7 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.3 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.8 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.1 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 2.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.4 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.2 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 1.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 1.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 1.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.4 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 0.4 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.1 | 1.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.9 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 1.5 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.2 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |