Project
DANIO-CODE
Navigation
Downloads
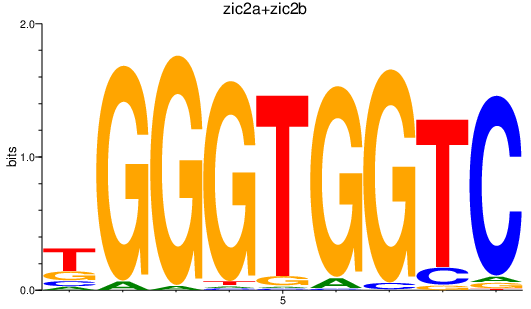
Results for zic2a+zic2b
Z-value: 1.33
Transcription factors associated with zic2a+zic2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zic2a
|
ENSDARG00000015554 | zic family member 2 (odd-paired homolog, Drosophila), a |
|
zic2b
|
ENSDARG00000037178 | zic family member 2 (odd-paired homolog, Drosophila) b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zic2a | dr10_dc_chr9_+_31471391_31471461 | 0.81 | 1.3e-04 | Click! |
| zic2b | dr10_dc_chr1_+_29054033_29054118 | 0.67 | 4.1e-03 | Click! |
Activity profile of zic2a+zic2b motif
Sorted Z-values of zic2a+zic2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zic2a+zic2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_10919978 | 5.68 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr15_+_29153215 | 5.48 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr23_+_20496283 | 5.29 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr8_+_29953399 | 3.85 |
ENSDART00000149372
ENSDART00000007640 |
ptch1
ptch1
|
patched 1 patched 1 |
| chr12_+_42281025 | 3.81 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr16_-_14506846 | 3.80 |
ENSDART00000170957
|
crabp2a
|
cellular retinoic acid binding protein 2, a |
| chr9_-_8752098 | 3.77 |
|
|
|
| chr2_-_34010299 | 3.60 |
ENSDART00000140910
|
ptch2
|
patched 2 |
| chr18_+_23004984 | 3.57 |
ENSDART00000171871
|
cbfb
|
core-binding factor, beta subunit |
| chr23_-_27226280 | 3.42 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr17_+_45931793 | 3.38 |
ENSDART00000155372
|
kif26ab
|
kinesin family member 26Ab |
| chr1_-_19152361 | 3.11 |
ENSDART00000132958
|
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr6_-_39661423 | 3.08 |
ENSDART00000110380
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr7_-_69639922 | 3.07 |
|
|
|
| chr17_-_45383925 | 3.05 |
|
|
|
| chr3_-_55956030 | 2.78 |
ENSDART00000158163
|
srl
|
sarcalumenin |
| chr11_-_4216204 | 2.73 |
ENSDART00000121716
|
ENSDARG00000086300
|
ENSDARG00000086300 |
| chr16_-_46605991 | 2.67 |
ENSDART00000115248
|
zgc:174938
|
zgc:174938 |
| chr24_-_2278409 | 2.64 |
|
|
|
| chr3_+_56008829 | 2.61 |
|
|
|
| chr10_+_32160464 | 2.60 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr14_+_25168063 | 2.60 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr5_+_26840147 | 2.59 |
ENSDART00000064701
|
loxl2b
|
lysyl oxidase-like 2b |
| chr23_-_27226387 | 2.44 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr9_+_8752130 | 2.41 |
ENSDART00000137813
|
FP103004.1
|
ENSDARG00000092413 |
| chr19_-_18316262 | 2.41 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr8_-_47811709 | 2.36 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr6_-_59510397 | 2.31 |
ENSDART00000170685
|
gli1
|
GLI family zinc finger 1 |
| chr17_+_27417635 | 2.30 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr9_-_46272322 | 2.27 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr13_-_39821399 | 2.24 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr20_+_15653121 | 2.21 |
ENSDART00000152702
|
jun
|
jun proto-oncogene |
| chr18_+_23891068 | 2.20 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr12_-_17357133 | 2.18 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr21_-_30378764 | 2.15 |
ENSDART00000136247
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr21_-_38534139 | 2.14 |
ENSDART00000148939
|
pof1b
|
premature ovarian failure, 1B |
| chr11_-_18542803 | 2.14 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr9_-_23081250 | 2.14 |
|
|
|
| chr17_-_6191539 | 2.10 |
ENSDART00000081707
|
ENSDARG00000058775
|
ENSDARG00000058775 |
| chr12_+_27026112 | 1.99 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr14_-_30363703 | 1.98 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin-like extracellular matrix protein 2a |
| chr23_-_9556986 | 1.96 |
ENSDART00000139688
|
soga1
|
suppressor of glucose, autophagy associated 1 |
| chr17_-_35004687 | 1.93 |
|
|
|
| chr16_+_1338291 | 1.88 |
ENSDART00000149299
|
cers2b
|
ceramide synthase 2b |
| chr10_-_15090647 | 1.88 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr23_+_34079219 | 1.87 |
ENSDART00000132668
|
si:ch211-207e14.4
|
si:ch211-207e14.4 |
| chr5_+_44316830 | 1.85 |
|
|
|
| chr16_+_32605735 | 1.82 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr10_+_43360137 | 1.79 |
ENSDART00000012522
|
vcanb
|
versican b |
| chr3_+_17210396 | 1.79 |
ENSDART00000090676
|
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr3_+_13114740 | 1.73 |
ENSDART00000162724
|
ENSDARG00000074231
|
ENSDARG00000074231 |
| chr4_+_22020246 | 1.73 |
ENSDART00000112035
ENSDART00000127664 |
myf5
|
myogenic factor 5 |
| chr5_+_32732535 | 1.71 |
ENSDART00000156913
|
BX927184.1
|
ENSDARG00000097904 |
| chr20_-_28739099 | 1.69 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr18_+_23891152 | 1.69 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr21_-_22914947 | 1.65 |
ENSDART00000083449
|
dub
|
duboraya |
| chr13_+_41998500 | 1.61 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr6_+_9192009 | 1.58 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr25_+_1773157 | 1.58 |
|
|
|
| chr6_+_36965023 | 1.56 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr9_-_20562293 | 1.55 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr5_+_37363015 | 1.55 |
ENSDART00000051224
|
pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr19_+_15538967 | 1.52 |
ENSDART00000171403
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr3_-_19942214 | 1.49 |
ENSDART00000029386
|
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr17_+_45931756 | 1.46 |
ENSDART00000155372
|
kif26ab
|
kinesin family member 26Ab |
| chr18_+_20504980 | 1.41 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| KN150339v1_-_34304 | 1.39 |
|
|
|
| chr14_+_34174018 | 1.34 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr5_+_40724281 | 1.31 |
ENSDART00000134112
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr24_-_6048914 | 1.30 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr19_-_18316039 | 1.29 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr24_-_16995194 | 1.29 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr17_+_45932080 | 1.26 |
ENSDART00000157326
|
kif26ab
|
kinesin family member 26Ab |
| chr14_+_31564252 | 1.26 |
|
|
|
| chr4_+_5124186 | 1.25 |
ENSDART00000127874
|
ccnd2b
|
cyclin D2, b |
| chr8_+_48488009 | 1.25 |
|
|
|
| chr6_+_7395817 | 1.24 |
ENSDART00000150939
ENSDART00000151114 |
myh10
|
myosin, heavy chain 10, non-muscle |
| chr23_+_22051774 | 1.24 |
ENSDART00000145172
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr18_+_23890892 | 1.23 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr22_+_38220826 | 1.23 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr19_+_43908885 | 1.22 |
ENSDART00000159421
|
BX649384.3
|
ENSDARG00000100839 |
| chr14_+_24637864 | 1.21 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr24_-_16994956 | 1.19 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr19_-_22983970 | 1.17 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr24_+_252774 | 1.16 |
ENSDART00000178656
ENSDART00000175867 |
CABZ01080224.1
|
ENSDARG00000107432 |
| chr14_-_10065562 | 1.15 |
ENSDART00000003879
|
fam199x
|
family with sequence similarity 199, X-linked |
| chr13_+_24148474 | 1.11 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr23_-_31585883 | 1.06 |
ENSDART00000157511
|
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr21_-_44707326 | 1.05 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr20_+_22780756 | 1.00 |
|
|
|
| chr3_-_62143464 | 0.98 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr3_+_23538277 | 0.98 |
ENSDART00000111227
|
hoxb10a
|
homeo box B10a |
| chr9_-_8752195 | 0.95 |
|
|
|
| chr8_+_29733109 | 0.94 |
ENSDART00000020621
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr16_-_7963894 | 0.94 |
ENSDART00000172836
|
CABZ01061349.1
|
ENSDARG00000105487 |
| chr7_-_34854886 | 0.91 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr19_+_24398194 | 0.89 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr23_-_7283514 | 0.88 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr11_+_25248195 | 0.88 |
ENSDART00000026249
|
gnl3l
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr22_-_14103803 | 0.87 |
ENSDART00000062902
|
ENSDARG00000042857
|
ENSDARG00000042857 |
| chr6_+_4000287 | 0.87 |
ENSDART00000111817
|
trim25l
|
tripartite motif containing 25, like |
| chr15_-_26911132 | 0.86 |
ENSDART00000077582
|
pitpnm3
|
PITPNM family member 3 |
| chr8_+_7297981 | 0.86 |
ENSDART00000170975
|
BX469901.2
|
ENSDARG00000103232 |
| chr18_+_6418004 | 0.86 |
ENSDART00000169401
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr11_-_36088889 | 0.84 |
ENSDART00000146495
|
psma5
|
proteasome subunit alpha 5 |
| chr11_-_25222937 | 0.83 |
|
|
|
| chr5_+_45823481 | 0.82 |
|
|
|
| chr3_+_24007046 | 0.79 |
|
|
|
| chr21_-_44707514 | 0.79 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr19_-_22984081 | 0.79 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr2_+_27000558 | 0.78 |
ENSDART00000099208
|
asph
|
aspartate beta-hydroxylase |
| chr9_+_14180954 | 0.78 |
ENSDART00000148055
|
si:ch211-67e16.11
|
si:ch211-67e16.11 |
| chr6_-_6816548 | 0.77 |
|
|
|
| chr1_-_21241192 | 0.75 |
|
|
|
| chr7_+_25735562 | 0.73 |
ENSDART00000112908
|
pelp1
|
proline, glutamate and leucine rich protein 1 |
| chr17_-_120568 | 0.73 |
|
|
|
| chr6_+_10491884 | 0.71 |
ENSDART00000151017
|
BX936397.2
|
ENSDARG00000096373 |
| chr13_-_37320497 | 0.70 |
|
|
|
| chr2_-_844040 | 0.69 |
ENSDART00000028159
|
foxf2a
|
forkhead box F2a |
| chr3_-_55278137 | 0.66 |
ENSDART00000123544
|
tex2
|
testis expressed 2 |
| chr5_-_52197945 | 0.66 |
|
|
|
| chr20_+_26196386 | 0.65 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr18_+_20504641 | 0.64 |
|
|
|
| chr16_-_22260272 | 0.63 |
ENSDART00000138419
|
CR855277.1
|
ENSDARG00000092020 |
| chr20_+_25727285 | 0.62 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr1_-_18880244 | 0.61 |
ENSDART00000131614
|
si:dkey-245p14.7
|
si:dkey-245p14.7 |
| chr3_+_56507117 | 0.61 |
ENSDART00000154405
|
rac1b
|
ras-related C3 botulinum toxin substrate 1b (rho family, small GTP binding protein Rac1) |
| chr19_-_20148533 | 0.59 |
ENSDART00000163790
|
evx1
|
even-skipped homeobox 1 |
| chr3_-_19942332 | 0.59 |
ENSDART00000124326
|
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr18_+_49230636 | 0.58 |
ENSDART00000167609
ENSDART00000135026 ENSDART00000171618 |
si:ch211-136a13.1
|
si:ch211-136a13.1 |
| chr20_-_25727347 | 0.58 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr22_-_14103942 | 0.58 |
ENSDART00000140323
|
ENSDARG00000042857
|
ENSDARG00000042857 |
| chr21_+_11686197 | 0.55 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr10_-_13158018 | 0.55 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr19_+_15538619 | 0.53 |
ENSDART00000151732
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr3_+_43125015 | 0.53 |
ENSDART00000165145
|
litaf
|
lipopolysaccharide-induced TNF factor |
| chr10_+_39325810 | 0.52 |
ENSDART00000170079
|
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr16_-_8606700 | 0.51 |
ENSDART00000137365
|
grb10b
|
growth factor receptor-bound protein 10b |
| chr20_+_32646617 | 0.51 |
|
|
|
| chr14_+_31564287 | 0.50 |
|
|
|
| chr2_-_59006559 | 0.50 |
|
|
|
| chr16_-_14506818 | 0.50 |
ENSDART00000170957
|
crabp2a
|
cellular retinoic acid binding protein 2, a |
| chr17_-_120755 | 0.49 |
|
|
|
| chr13_-_4238224 | 0.48 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr10_-_13438574 | 0.48 |
ENSDART00000030976
|
il11ra
|
interleukin 11 receptor, alpha |
| chr4_+_10367008 | 0.47 |
ENSDART00000138890
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr22_+_22694622 | 0.46 |
|
|
|
| chr13_+_41998172 | 0.45 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr10_+_17490443 | 0.45 |
|
|
|
| chr13_+_41998364 | 0.44 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr5_+_32732654 | 0.43 |
ENSDART00000156913
|
BX927184.1
|
ENSDARG00000097904 |
| chr14_-_50657786 | 0.43 |
ENSDART00000169456
|
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr15_+_509915 | 0.43 |
|
|
|
| chr6_+_40924749 | 0.41 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr7_+_37105966 | 0.40 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr5_+_35839198 | 0.40 |
ENSDART00000102973
ENSDART00000103020 |
eda
|
ectodysplasin A |
| chr5_-_29935062 | 0.38 |
ENSDART00000040328
|
h2afx
|
H2A histone family, member X |
| chr19_-_35688014 | 0.38 |
|
|
|
| chr13_-_4238408 | 0.37 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr16_-_42235969 | 0.36 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr8_+_31110356 | 0.36 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr3_-_55278055 | 0.36 |
ENSDART00000123544
|
tex2
|
testis expressed 2 |
| chr10_+_36096599 | 0.36 |
|
|
|
| chr4_+_43151489 | 0.35 |
ENSDART00000166967
|
si:ch211-226o13.2
|
si:ch211-226o13.2 |
| chr21_-_37914590 | 0.35 |
|
|
|
| chr11_+_2936759 | 0.31 |
ENSDART00000126071
|
cpne5b
|
copine Vb |
| chr5_-_14990023 | 0.28 |
|
|
|
| chr9_+_24344001 | 0.26 |
|
|
|
| chr5_-_56874791 | 0.26 |
|
|
|
| chr13_-_36785295 | 0.24 |
ENSDART00000167154
|
trim9
|
tripartite motif containing 9 |
| chr15_+_855739 | 0.22 |
ENSDART00000046783
|
zgc:113294
|
zgc:113294 |
| chr24_+_39405332 | 0.22 |
|
|
|
| chr7_+_20008453 | 0.22 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr20_-_51373292 | 0.21 |
ENSDART00000027836
ENSDART00000114407 |
rbm25b
|
RNA binding motif protein 25b |
| chr5_-_13335073 | 0.21 |
ENSDART00000114841
|
add2
|
adducin 2 (beta) |
| chr5_-_12789388 | 0.16 |
|
|
|
| chr7_+_25735735 | 0.14 |
ENSDART00000112908
|
pelp1
|
proline, glutamate and leucine rich protein 1 |
| chr3_-_40385511 | 0.13 |
|
|
|
| chr24_+_9371496 | 0.13 |
|
|
|
| chr19_-_10277398 | 0.13 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr11_+_34659252 | 0.13 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr3_+_32499605 | 0.13 |
ENSDART00000129339
|
srrm2
|
serine/arginine repetitive matrix 2 |
| chr8_-_49315811 | 0.11 |
ENSDART00000147020
|
prickle3
|
prickle homolog 3 |
| chr3_-_19942415 | 0.10 |
ENSDART00000124326
|
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr1_-_686007 | 0.10 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr17_-_747292 | 0.09 |
ENSDART00000159458
|
zbtb42
|
zinc finger and BTB domain containing 42 |
| chr15_-_24243608 | 0.07 |
ENSDART00000077980
|
pipox
|
pipecolic acid oxidase |
| chr11_+_18862603 | 0.04 |
|
|
|
| chr19_-_22984023 | 0.04 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr5_-_23296575 | 0.03 |
ENSDART00000134184
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr25_+_29631947 | 0.03 |
ENSDART00000154849
|
ENSDARG00000029431
|
ENSDARG00000029431 |
| chr1_-_58183299 | 0.01 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.5 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.9 | 3.6 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.9 | 2.6 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.8 | 2.3 | GO:0021742 | glossopharyngeal nerve development(GO:0021563) abducens nucleus development(GO:0021742) |
| 0.8 | 2.3 | GO:0036073 | perichondral ossification(GO:0036073) |
| 0.7 | 2.2 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.4 | 1.8 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.4 | 1.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.4 | 1.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.4 | 3.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.4 | 2.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.4 | 2.2 | GO:0000103 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.3 | 5.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 5.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.3 | 0.8 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.9 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.2 | 4.3 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.2 | 1.1 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 2.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 1.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 2.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.6 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.5 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 1.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 1.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 2.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.0 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 2.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 1.8 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 2.6 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 1.4 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.6 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.5 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 1.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 6.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 1.6 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.4 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 1.8 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 2.0 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.1 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:0019477 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 1.5 | GO:0016042 | lipid catabolic process(GO:0016042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.6 | 2.3 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.5 | 2.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 1.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 5.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 0.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 5.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 6.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.6 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 5.6 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 1.1 | 5.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 1.0 | 3.1 | GO:0008465 | glycerate dehydrogenase activity(GO:0008465) |
| 0.7 | 3.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.5 | 1.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 2.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.4 | 1.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.4 | 4.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 2.2 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 5.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 0.6 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.2 | 1.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 2.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 2.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 2.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 1.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 2.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 6.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.5 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 1.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.9 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.7 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.0 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.6 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.8 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 2.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 1.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 3.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 1.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 7.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 1.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |