Project
DANIO-CODE
Navigation
Downloads
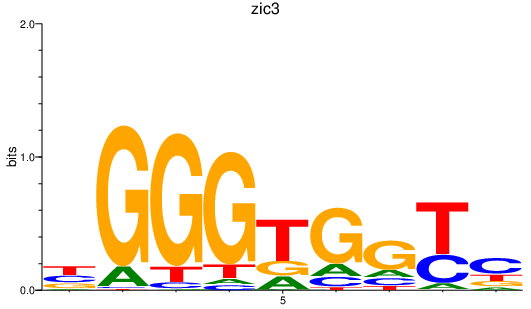
Results for zic3
Z-value: 0.43
Transcription factors associated with zic3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zic3
|
ENSDARG00000071497 | zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zic3 | dr10_dc_chr14_-_31676235_31676332 | -0.88 | 5.6e-06 | Click! |
Activity profile of zic3 motif
Sorted Z-values of zic3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zic3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_20236308 | 0.96 |
|
|
|
| chr22_+_37688839 | 0.91 |
ENSDART00000174664
ENSDART00000178200 |
FO704589.1
|
ENSDARG00000108910 |
| chr19_+_24398149 | 0.79 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr9_-_3548211 | 0.74 |
ENSDART00000145043
ENSDART00000169586 |
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr6_+_27100350 | 0.69 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr22_+_37689055 | 0.60 |
|
|
|
| chr6_+_27100249 | 0.51 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr9_-_9729041 | 0.51 |
ENSDART00000175212
|
gpr156
|
G protein-coupled receptor 156 |
| chr2_-_30800968 | 0.45 |
ENSDART00000056735
|
rgs20
|
regulator of G protein signaling 20 |
| chr25_-_34235582 | 0.39 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr22_+_37688988 | 0.37 |
|
|
|
| chr25_-_34235633 | 0.37 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr2_-_7772932 | 0.37 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr1_-_39141284 | 0.36 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr22_-_37630428 | 0.21 |
ENSDART00000149482
|
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr7_-_44455032 | 0.15 |
ENSDART00000019270
|
nae1
|
nedd8 activating enzyme E1 subunit 1 |
| chr16_-_20044830 | 0.05 |
|
|
|
| chr11_+_34659252 | 0.05 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046078 | dUMP biosynthetic process(GO:0006226) dUMP metabolic process(GO:0046078) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |