Project
DANIO-CODE
Navigation
Downloads
Results for zic4+zic6
Z-value: 0.36
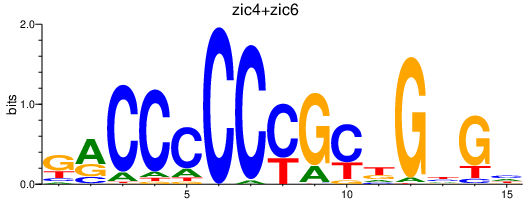
Transcription factors associated with zic4+zic6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zic4
|
ENSDARG00000031307 | zic family member 4 |
|
zic6
|
ENSDARG00000071496 | zic family member 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zic6 | dr10_dc_chr14_+_31682159_31682218 | 0.58 | 1.9e-02 | Click! |
| zic4 | dr10_dc_chr24_+_4946542_4946628 | 0.53 | 3.3e-02 | Click! |
Activity profile of zic4+zic6 motif
Sorted Z-values of zic4+zic6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of zic4+zic6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_32007035 | 1.05 |
ENSDART00000132981
|
si:dkey-126g1.9
|
si:dkey-126g1.9 |
| chr7_-_32752138 | 0.92 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr13_-_39821399 | 0.84 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr15_+_37039861 | 0.80 |
ENSDART00000172664
|
kirrel3l
|
kin of IRRE like 3 like |
| chr3_-_23558171 | 0.77 |
|
|
|
| chr23_-_5785468 | 0.74 |
ENSDART00000033093
|
lad1
|
ladinin |
| chr2_+_25363717 | 0.71 |
ENSDART00000142601
|
stag1a
|
stromal antigen 1a |
| chr22_+_17091411 | 0.70 |
ENSDART00000170076
|
frem1b
|
Fras1 related extracellular matrix 1b |
| chr18_-_46356213 | 0.63 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr20_-_9992898 | 0.54 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr3_-_39313029 | 0.53 |
ENSDART00000156123
|
CU464118.1
|
ENSDARG00000097747 |
| chr12_-_26760324 | 0.51 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr5_+_44407763 | 0.49 |
ENSDART00000010786
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr25_-_352985 | 0.49 |
ENSDART00000165705
|
szl
|
sizzled |
| chr25_-_13631707 | 0.42 |
ENSDART00000169865
|
lcat
|
lecithin-cholesterol acyltransferase |
| chr8_+_54171992 | 0.41 |
ENSDART00000122692
|
CABZ01112317.1
|
ENSDARG00000086057 |
| chr23_-_28367816 | 0.41 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr20_-_23184142 | 0.40 |
ENSDART00000176282
|
CU693368.2
|
ENSDARG00000108718 |
| KN149968v1_+_15749 | 0.39 |
ENSDART00000168977
|
ENSDARG00000102503
|
ENSDARG00000102503 |
| chr2_-_53772219 | 0.39 |
ENSDART00000037557
|
admp
|
anti-dorsalizing morphogenic protein |
| chr1_-_8968543 | 0.38 |
ENSDART00000126877
ENSDART00000123773 ENSDART00000126996 |
ugt5b1
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B1 UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr6_-_7281412 | 0.38 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr8_+_6989445 | 0.38 |
ENSDART00000134440
|
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr23_+_32007004 | 0.38 |
ENSDART00000145639
|
si:dkey-126g1.9
|
si:dkey-126g1.9 |
| chr7_+_73548430 | 0.34 |
ENSDART00000092691
|
pea15
|
phosphoprotein enriched in astrocytes 15 |
| chr1_-_45709828 | 0.33 |
ENSDART00000150029
|
atp11a
|
ATPase, class VI, type 11A |
| chr2_+_15459785 | 0.33 |
ENSDART00000016071
ENSDART00000141921 |
arhgap29b
|
Rho GTPase activating protein 29b |
| chr16_-_55320569 | 0.32 |
ENSDART00000156368
|
ENSDARG00000069583
|
ENSDARG00000069583 |
| chr13_-_32517896 | 0.30 |
ENSDART00000132820
|
bend3
|
BEN domain containing 3 |
| chr23_+_7537089 | 0.29 |
ENSDART00000135551
|
si:ch211-200e2.1
|
si:ch211-200e2.1 |
| chr7_+_30516734 | 0.28 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr5_+_45823481 | 0.27 |
|
|
|
| chr23_-_27621359 | 0.23 |
ENSDART00000144419
|
larp4aa
|
La ribonucleoprotein domain family, member 4Aa |
| chr18_+_20058028 | 0.23 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr6_-_16156494 | 0.21 |
|
|
|
| chr14_-_32163041 | 0.21 |
ENSDART00000034883
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr2_-_58822653 | 0.20 |
ENSDART00000114286
ENSDART00000114181 |
sugp1
|
SURP and G patch domain containing 1 |
| chr25_-_13737344 | 0.19 |
|
|
|
| chr20_-_28739099 | 0.18 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr2_+_42342148 | 0.18 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr3_+_30962542 | 0.17 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr22_-_6287015 | 0.16 |
ENSDART00000148832
|
AL928685.4
|
ENSDARG00000095907 |
| chr10_+_17490443 | 0.16 |
|
|
|
| chr6_-_7281346 | 0.16 |
ENSDART00000133096
|
fkbp11
|
FK506 binding protein 11 |
| chr22_+_29694605 | 0.15 |
|
|
|
| chr12_-_41124276 | 0.15 |
ENSDART00000172022
ENSDART00000158605 ENSDART00000162967 |
dpysl4
|
dihydropyrimidinase-like 4 |
| chr23_-_36319185 | 0.15 |
ENSDART00000139328
|
znf740b
|
zinc finger protein 740b |
| chr10_-_44675914 | 0.15 |
|
|
|
| chr24_-_34794463 | 0.15 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr8_-_7812788 | 0.15 |
ENSDART00000160692
|
zgc:113363
|
zgc:113363 |
| chr23_+_44249976 | 0.14 |
ENSDART00000158305
|
tec
|
tec protein tyrosine kinase |
| chr7_+_73548348 | 0.14 |
ENSDART00000092691
|
pea15
|
phosphoprotein enriched in astrocytes 15 |
| chr8_-_17031599 | 0.14 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr16_-_49672970 | 0.13 |
ENSDART00000127513
|
znf385d
|
zinc finger protein 385D |
| chr3_-_40020749 | 0.13 |
ENSDART00000129664
|
drg2
|
developmentally regulated GTP binding protein 2 |
| chr24_-_25545773 | 0.12 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr1_+_11541423 | 0.11 |
ENSDART00000170760
|
tmod1
|
tropomodulin 1 |
| chr6_+_17928720 | 0.11 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr13_+_37527958 | 0.11 |
ENSDART00000141988
|
phf3
|
PHD finger protein 3 |
| chr16_-_25793780 | 0.10 |
|
|
|
| chr5_+_68925227 | 0.09 |
|
|
|
| chr15_+_37040064 | 0.09 |
ENSDART00000172664
|
kirrel3l
|
kin of IRRE like 3 like |
| chr6_+_6640324 | 0.08 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr6_-_16156294 | 0.08 |
ENSDART00000160809
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr3_-_40020899 | 0.08 |
ENSDART00000025285
|
drg2
|
developmentally regulated GTP binding protein 2 |
| chr14_+_40485238 | 0.06 |
ENSDART00000128588
|
taf7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr12_-_13611954 | 0.06 |
ENSDART00000124638
ENSDART00000124364 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr3_+_16880842 | 0.05 |
ENSDART00000080854
|
stat3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr18_+_30443201 | 0.05 |
ENSDART00000140908
|
gse1
|
Gse1 coiled-coil protein |
| KN149739v1_+_40107 | 0.05 |
|
|
|
| chr11_+_31295037 | 0.04 |
ENSDART00000140204
|
egln1b
|
egl-9 family hypoxia-inducible factor 1b |
| chr8_-_4562273 | 0.04 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| KN150222v1_+_74557 | 0.03 |
|
|
|
| chr25_-_5805538 | 0.03 |
|
|
|
| chr18_+_49230636 | 0.02 |
ENSDART00000167609
ENSDART00000135026 ENSDART00000171618 |
si:ch211-136a13.1
|
si:ch211-136a13.1 |
| chr3_+_23964015 | 0.01 |
ENSDART00000138270
|
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr18_-_46355957 | 0.01 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr23_+_24778090 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.2 | 0.5 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.5 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.4 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.4 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.1 | 0.9 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.5 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.7 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |