Project
DANIO-CODE
Navigation
Downloads
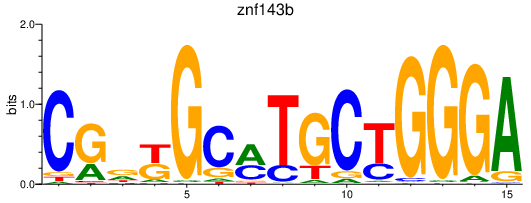
Results for znf143b
Z-value: 1.12
Transcription factors associated with znf143b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf143b
|
ENSDARG00000041581 | zinc finger protein 143b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf143b | dr10_dc_chr18_-_16948109_16948125 | 0.68 | 3.4e-03 | Click! |
Activity profile of znf143b motif
Sorted Z-values of znf143b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of znf143b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_43641419 | 2.79 |
ENSDART00000151486
ENSDART00000151115 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr7_-_47978449 | 2.68 |
ENSDART00000127007
ENSDART00000024062 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr11_+_29299382 | 2.40 |
|
|
|
| chr6_-_50731449 | 2.24 |
ENSDART00000157153
|
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr4_+_1770595 | 2.21 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr15_-_9443212 | 2.11 |
ENSDART00000177158
|
sacs
|
sacsin molecular chaperone |
| chr18_+_5616001 | 2.05 |
ENSDART00000163629
|
dut
|
deoxyuridine triphosphatase |
| chr4_+_4825461 | 2.04 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr18_+_42925672 | 1.99 |
|
|
|
| chr12_+_48590478 | 1.95 |
|
|
|
| chr3_+_17901295 | 1.90 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr2_-_34154845 | 1.89 |
ENSDART00000133381
|
cenpl
|
centromere protein L |
| chr3_+_1451017 | 1.83 |
ENSDART00000161945
|
rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr16_+_42567707 | 1.79 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr18_-_8422329 | 1.75 |
ENSDART00000081143
|
sephs1
|
selenophosphate synthetase 1 |
| chr16_+_19831573 | 1.71 |
ENSDART00000135359
|
macc1
|
metastasis associated in colon cancer 1 |
| chr16_+_28587065 | 1.46 |
|
|
|
| chr21_-_11562388 | 1.45 |
ENSDART00000139289
|
cast
|
calpastatin |
| chr14_-_6901209 | 1.44 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr11_+_26896056 | 1.43 |
ENSDART00000158411
ENSDART00000173374 ENSDART00000129736 |
hdac11
|
histone deacetylase 11 |
| chr20_+_43794079 | 1.39 |
ENSDART00000045185
|
lin9
|
lin-9 DREAM MuvB core complex component |
| chr18_-_199009 | 1.38 |
|
|
|
| chr22_-_5888816 | 1.38 |
ENSDART00000141373
|
si:rp71-36a1.1
|
si:rp71-36a1.1 |
| chr4_+_14983045 | 1.37 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr18_+_18011759 | 1.37 |
ENSDART00000147797
|
ENSDARG00000011498
|
ENSDARG00000011498 |
| chr12_-_998693 | 1.36 |
ENSDART00000152346
|
polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr13_+_15685745 | 1.34 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr7_-_55879342 | 1.33 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr15_+_5124690 | 1.33 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr15_+_5124897 | 1.32 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr15_+_44160948 | 1.29 |
ENSDART00000110060
|
zgc:165514
|
zgc:165514 |
| chr15_+_24741931 | 1.28 |
ENSDART00000143137
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr5_+_33949157 | 1.28 |
ENSDART00000136787
|
aif1l
|
allograft inflammatory factor 1-like |
| chr1_+_49263896 | 1.27 |
|
|
|
| chr4_+_1770712 | 1.25 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| KN150435v1_-_98375 | 1.24 |
ENSDART00000164042
|
klc2
|
kinesin light chain 2 |
| chr8_+_19945528 | 1.23 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr11_+_26895952 | 1.21 |
ENSDART00000158411
ENSDART00000173374 ENSDART00000129736 |
hdac11
|
histone deacetylase 11 |
| chr15_-_1657417 | 1.21 |
ENSDART00000034456
|
kpna4
|
karyopherin alpha 4 (importin alpha 3) |
| chr14_+_32512043 | 1.21 |
ENSDART00000166351
|
nkrf
|
NFKB repressing factor |
| chr8_+_19945820 | 1.20 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr3_+_17901095 | 1.18 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr3_+_17900981 | 1.18 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr5_+_33949689 | 1.17 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr15_+_24741620 | 1.14 |
ENSDART00000078014
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr20_-_16949608 | 1.14 |
ENSDART00000027582
|
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr20_+_15653221 | 1.13 |
ENSDART00000152167
|
jun
|
jun proto-oncogene |
| chr14_-_30536363 | 1.12 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr17_-_6225136 | 1.10 |
ENSDART00000137389
ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr5_-_12853593 | 1.09 |
ENSDART00000099602
|
ube2l3b
|
ubiquitin-conjugating enzyme E2L 3b |
| chr15_-_6969345 | 1.09 |
ENSDART00000169529
|
mrps22
|
mitochondrial ribosomal protein S22 |
| chr21_+_7570595 | 1.08 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr2_+_31454465 | 1.07 |
ENSDART00000176837
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr12_-_46943717 | 1.07 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr5_-_23079460 | 1.06 |
ENSDART00000027217
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr3_-_33296077 | 1.05 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr4_+_4825409 | 1.04 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr2_+_31454204 | 1.03 |
ENSDART00000176837
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr16_-_39317068 | 1.01 |
ENSDART00000133642
|
gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr16_+_28587293 | 1.00 |
|
|
|
| chr17_+_46403915 | 1.00 |
ENSDART00000154499
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr20_+_44600015 | 1.00 |
ENSDART00000023763
|
wdcp
|
WD repeat and coiled coil containing |
| chr11_-_1939192 | 0.99 |
ENSDART00000172885
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr12_+_48590522 | 0.98 |
|
|
|
| KN149861v1_-_8998 | 0.97 |
|
|
|
| chr5_+_33949469 | 0.97 |
ENSDART00000136787
|
aif1l
|
allograft inflammatory factor 1-like |
| chr23_+_7758140 | 0.96 |
ENSDART00000164255
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr8_-_22537415 | 0.96 |
ENSDART00000165640
|
porcnl
|
porcupine homolog like |
| chr16_-_23975853 | 0.96 |
ENSDART00000077807
|
BX004983.1
|
ENSDARG00000055446 |
| chr7_-_11363817 | 0.95 |
ENSDART00000174163
|
CABZ01068273.1
|
ENSDARG00000105718 |
| chr8_+_29733037 | 0.95 |
ENSDART00000133955
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr15_-_56306 | 0.95 |
ENSDART00000168340
|
CABZ01080370.1
|
POU class 2 homeobox 1a |
| chr11_+_5883229 | 0.94 |
ENSDART00000129663
|
dazap1
|
DAZ associated protein 1 |
| chr12_+_19999188 | 0.93 |
|
|
|
| chr9_-_30174641 | 0.93 |
ENSDART00000134157
ENSDART00000089206 |
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr3_-_40060674 | 0.92 |
ENSDART00000175118
|
znf598
|
zinc finger protein 598 |
| chr7_+_67525979 | 0.92 |
ENSDART00000162978
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr4_+_30530 | 0.91 |
ENSDART00000157825
|
syn3
|
synapsin III |
| chr22_+_17506147 | 0.91 |
ENSDART00000142871
|
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr11_-_1061904 | 0.91 |
ENSDART00000173253
ENSDART00000172765 |
CU914760.1
|
ENSDARG00000105313 |
| chr10_-_35242344 | 0.90 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr2_+_34154885 | 0.90 |
ENSDART00000137170
|
dars2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr2_-_24909432 | 0.90 |
ENSDART00000170283
|
crsp7
|
cofactor required for Sp1 transcriptional activation, subunit 7 |
| chr21_+_13689239 | 0.89 |
ENSDART00000112451
|
slc31a2
|
solute carrier family 31 (copper transporter), member 2 |
| chr2_-_24909546 | 0.88 |
ENSDART00000170283
|
crsp7
|
cofactor required for Sp1 transcriptional activation, subunit 7 |
| chr8_-_43151763 | 0.88 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr21_+_7868319 | 0.87 |
ENSDART00000121813
ENSDART00000021272 |
wdr41
|
WD repeat domain 41 |
| chr15_+_25516764 | 0.87 |
ENSDART00000009545
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr1_+_54417692 | 0.86 |
ENSDART00000132727
|
aftpha
|
aftiphilin a |
| chr2_-_37555425 | 0.86 |
ENSDART00000143496
|
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr21_+_7868073 | 0.86 |
ENSDART00000121813
ENSDART00000021272 |
wdr41
|
WD repeat domain 41 |
| chr9_-_29769100 | 0.86 |
ENSDART00000140876
|
cenpj
|
centromere protein J |
| KN149861v1_-_8662 | 0.84 |
ENSDART00000179340
|
CABZ01089907.1
|
ENSDARG00000107716 |
| chr15_-_24741104 | 0.84 |
ENSDART00000100756
|
tmem199
|
transmembrane protein 199 |
| chr20_-_16949680 | 0.83 |
ENSDART00000027582
|
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr7_-_61693560 | 0.83 |
ENSDART00000062704
|
plaa
|
phospholipase A2-activating protein |
| chr2_+_55460609 | 0.82 |
ENSDART00000016143
|
ENSDARG00000001817
|
ENSDARG00000001817 |
| chr18_+_49974874 | 0.82 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr19_+_40482359 | 0.82 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr16_+_42567668 | 0.81 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr21_+_21754830 | 0.79 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr6_-_27067072 | 0.79 |
ENSDART00000149363
|
stk25a
|
serine/threonine kinase 25a |
| chr17_+_28689850 | 0.78 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr23_+_31319119 | 0.75 |
ENSDART00000133391
|
irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr4_+_11484516 | 0.75 |
ENSDART00000140954
|
asb13a.2
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 2 |
| chr14_+_41042379 | 0.75 |
ENSDART00000173335
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr3_-_34687314 | 0.74 |
ENSDART00000084448
|
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr4_-_14982671 | 0.72 |
|
|
|
| chr15_+_25516962 | 0.72 |
ENSDART00000165509
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr4_+_1770500 | 0.72 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr17_+_23450965 | 0.71 |
ENSDART00000154716
|
kif20ba
|
kinesin family member 20Ba |
| chr3_-_54591713 | 0.71 |
ENSDART00000074010
|
ubald1b
|
UBA-like domain containing 1b |
| chr23_+_44069409 | 0.69 |
ENSDART00000149088
|
nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| KN150435v1_-_98185 | 0.68 |
ENSDART00000164042
|
klc2
|
kinesin light chain 2 |
| chr15_+_33985567 | 0.68 |
ENSDART00000169037
|
inpp5ka
|
inositol polyphosphate-5-phosphatase Ka |
| chr21_-_23009910 | 0.68 |
ENSDART00000016167
|
zw10
|
zw10 kinetochore protein |
| chr20_+_14914870 | 0.68 |
ENSDART00000010550
|
pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr20_-_15262089 | 0.67 |
ENSDART00000063882
|
plpp6
|
phospholipid phosphatase 6 |
| chr14_-_6901783 | 0.67 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr4_+_4825628 | 0.66 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr8_-_51537679 | 0.65 |
ENSDART00000147742
|
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr25_-_20160885 | 0.62 |
ENSDART00000160700
|
dnm1l
|
dynamin 1-like |
| chr21_-_23873574 | 0.62 |
|
|
|
| chr23_+_17077597 | 0.62 |
|
|
|
| chr5_+_22632955 | 0.62 |
ENSDART00000178821
|
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr5_+_22632867 | 0.62 |
ENSDART00000178821
|
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr17_-_1949701 | 0.62 |
ENSDART00000013093
ENSDART00000168231 ENSDART00000166833 |
zfyve19
|
zinc finger, FYVE domain containing 19 |
| chr4_-_9780251 | 0.60 |
ENSDART00000080744
|
svopl
|
SVOP-like |
| chr10_+_44209117 | 0.60 |
|
|
|
| chr15_+_5124820 | 0.60 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr14_+_971438 | 0.59 |
ENSDART00000161487
|
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr2_-_27892824 | 0.59 |
|
|
|
| chr15_+_5124616 | 0.59 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr1_-_49307663 | 0.59 |
|
|
|
| chr11_+_1939273 | 0.59 |
ENSDART00000155304
|
CT573157.1
|
ENSDARG00000096842 |
| chr6_+_38775771 | 0.59 |
ENSDART00000129655
|
ube3a
|
ubiquitin protein ligase E3A |
| chr22_-_469154 | 0.58 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr25_+_8585585 | 0.58 |
ENSDART00000154680
|
man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr18_+_18011800 | 0.57 |
ENSDART00000147797
|
ENSDARG00000011498
|
ENSDARG00000011498 |
| chr1_+_49263812 | 0.57 |
|
|
|
| chr25_-_13774670 | 0.56 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr2_-_24909674 | 0.56 |
ENSDART00000170283
|
crsp7
|
cofactor required for Sp1 transcriptional activation, subunit 7 |
| chr11_+_5883270 | 0.55 |
ENSDART00000130768
|
dazap1
|
DAZ associated protein 1 |
| chr18_-_50617626 | 0.54 |
ENSDART00000172530
|
ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr18_-_5633564 | 0.54 |
|
|
|
| chr13_+_15685540 | 0.54 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr9_-_34145854 | 0.53 |
|
|
|
| chr6_-_13586850 | 0.53 |
ENSDART00000111102
|
chpfb
|
chondroitin polymerizing factor b |
| chr21_+_17265099 | 0.53 |
ENSDART00000145057
|
tsc1b
|
tuberous sclerosis 1b |
| chr3_-_49031058 | 0.52 |
ENSDART00000154561
|
mrpl12
|
mitochondrial ribosomal protein L12 |
| chr2_+_42223071 | 0.52 |
ENSDART00000134203
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr20_-_34868582 | 0.52 |
|
|
|
| chr23_+_31319064 | 0.51 |
ENSDART00000146510
|
irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr4_+_1770198 | 0.51 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr6_+_20595096 | 0.51 |
|
|
|
| chr23_-_3568175 | 0.50 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr3_+_54777037 | 0.49 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr16_-_8218930 | 0.47 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr13_-_42274444 | 0.47 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr11_+_2083270 | 0.47 |
ENSDART00000162167
|
smug1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr3_-_30730348 | 0.46 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr9_+_26275438 | 0.45 |
ENSDART00000142272
|
arglu1a
|
arginine and glutamate rich 1a |
| chr18_-_16134320 | 0.44 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr21_+_7570643 | 0.44 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr23_+_34021303 | 0.44 |
ENSDART00000136104
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr14_+_15982295 | 0.44 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr16_-_5243563 | 0.44 |
ENSDART00000060630
|
ttk
|
ttk protein kinase |
| chr24_+_35082208 | 0.43 |
ENSDART00000113014
|
rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr4_-_9779987 | 0.43 |
ENSDART00000134280
|
svopl
|
SVOP-like |
| chr18_+_8273087 | 0.42 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr3_+_32346154 | 0.40 |
ENSDART00000103246
|
scaf1
|
SR-related CTD-associated factor 1 |
| chr3_-_30927275 | 0.38 |
ENSDART00000112266
|
armc5
|
armadillo repeat containing 5 |
| chr24_-_31155315 | 0.37 |
ENSDART00000168398
|
hccsb
|
holocytochrome c synthase b |
| chr1_+_54417596 | 0.36 |
ENSDART00000132727
|
aftpha
|
aftiphilin a |
| chr16_-_44910578 | 0.36 |
ENSDART00000157375
|
arhgap33
|
Rho GTPase activating protein 33 |
| chr23_+_34021480 | 0.36 |
ENSDART00000136104
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr7_-_55879197 | 0.35 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr15_+_44161047 | 0.34 |
ENSDART00000110060
|
zgc:165514
|
zgc:165514 |
| chr14_-_31222400 | 0.33 |
ENSDART00000034979
|
mospd1
|
motile sperm domain containing 1 |
| chr1_+_50333212 | 0.32 |
ENSDART00000022290
|
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr5_-_23079490 | 0.31 |
ENSDART00000027217
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr25_-_8077833 | 0.31 |
ENSDART00000164846
ENSDART00000006579 |
saal1
|
serum amyloid A-like 1 |
| chr18_-_15360903 | 0.30 |
ENSDART00000019818
|
ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr21_+_29970800 | 0.30 |
ENSDART00000150071
|
ttc1
|
tetratricopeptide repeat domain 1 |
| chr10_+_37238831 | 0.28 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr10_-_27047692 | 0.28 |
ENSDART00000139942
ENSDART00000146983 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr6_+_39057509 | 0.27 |
|
|
|
| chr18_+_5616306 | 0.27 |
ENSDART00000163629
|
dut
|
deoxyuridine triphosphatase |
| chr3_-_60883616 | 0.27 |
ENSDART00000156978
|
aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr20_-_9440244 | 0.26 |
ENSDART00000025330
|
rdh14b
|
retinol dehydrogenase 14b (all-trans/9-cis/11-cis) |
| chr1_+_20870346 | 0.26 |
ENSDART00000158648
|
primpol
|
primase and polymerase (DNA-directed) |
| chr19_+_31944417 | 0.25 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr19_+_7839888 | 0.25 |
|
|
|
| chr15_+_24741845 | 0.25 |
ENSDART00000143137
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr18_-_16134258 | 0.25 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr18_+_413580 | 0.24 |
ENSDART00000160279
|
lsm14ab
|
LSM14A mRNA processing body assembly factor b |
| chr16_+_24045675 | 0.23 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr23_+_7758022 | 0.22 |
ENSDART00000164255
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr7_+_25765060 | 0.21 |
|
|
|
| chr14_-_4117151 | 0.21 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.8 | 2.3 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.7 | 2.7 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.4 | 2.7 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.3 | 0.9 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.3 | 0.9 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.3 | 0.8 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 0.8 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.3 | 2.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 1.0 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 3.4 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.2 | 1.7 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.2 | 1.0 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 0.6 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.2 | 0.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.2 | 1.0 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 0.9 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.2 | 3.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 1.5 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 1.9 | GO:0009070 | serine family amino acid biosynthetic process(GO:0009070) |
| 0.1 | 0.7 | GO:0045719 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.8 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.8 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.5 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 1.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 4.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.9 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 1.5 | GO:0048026 | positive regulation of RNA splicing(GO:0033120) positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.3 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.3 | GO:0043589 | positive regulation of phospholipase C activity(GO:0010863) skin morphogenesis(GO:0043589) regulation of phospholipase C activity(GO:1900274) |
| 0.1 | 1.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.3 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.4 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.4 | GO:1902534 | lysosomal microautophagy(GO:0016237) pexophagy(GO:0030242) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.6 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.2 | GO:0006546 | glycine metabolic process(GO:0006544) glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 3.8 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.1 | GO:0060911 | cardiac cell fate commitment(GO:0060911) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0031281 | positive regulation of cyclic nucleotide metabolic process(GO:0030801) positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) positive regulation of nucleotide biosynthetic process(GO:0030810) positive regulation of cyclase activity(GO:0031281) positive regulation of lyase activity(GO:0051349) positive regulation of purine nucleotide biosynthetic process(GO:1900373) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.0 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 1.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.9 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.6 | GO:0002573 | myeloid leukocyte differentiation(GO:0002573) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.7 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.9 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.6 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.4 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.3 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 0.9 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.8 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 1.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.8 | GO:0000209 | protein polyubiquitination(GO:0000209) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.3 | 2.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 2.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 2.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 2.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 1.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 1.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 3.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.8 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.4 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 1.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 4.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 2.1 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 1.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.1 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.7 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.8 | 2.3 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.3 | 1.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.3 | 0.9 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.3 | 2.7 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 1.2 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.2 | 0.8 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.2 | 1.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 3.8 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.2 | 1.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 1.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 0.9 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 0.7 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 1.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.8 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.8 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.5 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 2.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 3.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.6 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 2.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 1.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 3.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 0.9 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 2.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 3.0 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 2.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.2 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 2.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |