Project
DANIO-CODE
Navigation
Downloads
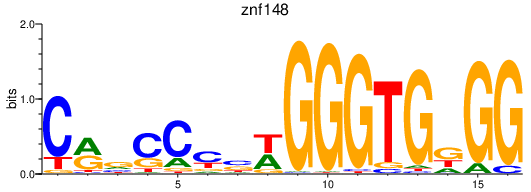
Results for znf148
Z-value: 0.42
Transcription factors associated with znf148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf148
|
ENSDARG00000055106 | zinc finger protein 148 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf148 | dr10_dc_chr9_+_38814714_38814787 | 0.16 | 5.5e-01 | Click! |
Activity profile of znf148 motif
Sorted Z-values of znf148 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of znf148
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_50452131 | 0.83 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr10_-_45155460 | 0.76 |
ENSDART00000170767
|
ved
|
ventrally expressed dharma/bozozok antagonist |
| chr11_-_18542803 | 0.73 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr21_-_11539798 | 0.70 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr6_+_36481951 | 0.67 |
ENSDART00000104248
|
lrrc15
|
leucine rich repeat containing 15 |
| chr6_-_9346394 | 0.48 |
ENSDART00000144335
|
cyp27c1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr20_+_17840364 | 0.43 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr13_-_36409205 | 0.41 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr5_+_6054781 | 0.38 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr16_+_40610589 | 0.35 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr6_+_28886741 | 0.33 |
ENSDART00000065137
|
tp63
|
tumor protein p63 |
| chr15_+_46114986 | 0.27 |
ENSDART00000156366
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr10_+_21477806 | 0.22 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr14_-_25879853 | 0.22 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr6_+_50452229 | 0.21 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr16_+_11138879 | 0.20 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr10_+_38568464 | 0.20 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr16_+_11138924 | 0.19 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr12_-_4497094 | 0.18 |
ENSDART00000163651
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr6_-_27963880 | 0.18 |
ENSDART00000163747
|
BX914220.1
|
ENSDARG00000098603 |
| chr10_+_187740 | 0.16 |
ENSDART00000167367
|
ets2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr6_-_8501230 | 0.16 |
ENSDART00000143956
|
cavin2b
|
caveolae associated protein 2b |
| KN149841v1_+_6400 | 0.13 |
ENSDART00000179534
|
CABZ01103873.1
|
ENSDARG00000108002 |
| chr5_+_41192815 | 0.13 |
ENSDART00000136439
|
bcl7a
|
B-cell CLL/lymphoma 7A |
| chr3_-_31792796 | 0.12 |
ENSDART00000122589
|
rnf113a
|
ring finger protein 113A |
| chr10_+_21477579 | 0.11 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr1_-_416138 | 0.11 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr22_-_570067 | 0.11 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr10_+_32007448 | 0.11 |
ENSDART00000019416
|
lhfp
|
lipoma HMGIC fusion partner |
| chr16_+_5778650 | 0.10 |
ENSDART00000131575
|
zgc:158689
|
zgc:158689 |
| chr6_+_50452529 | 0.10 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr16_-_33167598 | 0.10 |
|
|
|
| chr10_+_21477307 | 0.09 |
ENSDART00000064558
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr4_-_11752178 | 0.09 |
ENSDART00000102301
|
podxl
|
podocalyxin-like |
| chr10_-_2944221 | 0.09 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr12_-_4444385 | 0.08 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr7_-_26332236 | 0.08 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr5_+_25612590 | 0.07 |
ENSDART00000098514
|
oclnb
|
occludin b |
| chr20_+_38298755 | 0.07 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr16_-_25765396 | 0.07 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr13_+_42183490 | 0.07 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr15_+_42811722 | 0.06 |
ENSDART00000164879
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr7_-_26331734 | 0.06 |
|
|
|
| chr21_+_25199691 | 0.06 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr16_-_25765322 | 0.05 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr22_-_11408859 | 0.05 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr12_-_22549037 | 0.05 |
ENSDART00000164888
|
zcchc4
|
zinc finger, CCHC domain containing 4 |
| chr5_+_41193186 | 0.05 |
ENSDART00000136439
|
bcl7a
|
B-cell CLL/lymphoma 7A |
| chr14_-_2166955 | 0.05 |
ENSDART00000169201
|
pcdh2g16
|
protocadherin 2 gamma 16 |
| chr11_+_43823529 | 0.05 |
ENSDART00000130273
|
traf3ip2b
|
TRAF3 interacting protein 2b |
| chr5_-_20527105 | 0.05 |
ENSDART00000133461
|
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr5_-_27953681 | 0.04 |
ENSDART00000078708
|
zgc:162952
|
zgc:162952 |
| chr9_-_30463616 | 0.04 |
ENSDART00000089526
|
otc
|
ornithine carbamoyltransferase |
| chr4_-_11752109 | 0.04 |
ENSDART00000102301
|
podxl
|
podocalyxin-like |
| chr9_-_7560915 | 0.04 |
ENSDART00000081553
|
desma
|
desmin a |
| chr13_-_42453219 | 0.03 |
ENSDART00000135172
|
fam83ha
|
family with sequence similarity 83, member Ha |
| KN149841v1_+_6481 | 0.03 |
ENSDART00000179534
|
CABZ01103873.1
|
ENSDARG00000108002 |
| chr16_+_54729975 | 0.03 |
|
|
|
| chr24_-_40968409 | 0.03 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr17_-_52550596 | 0.02 |
ENSDART00000084268
|
fam98b
|
family with sequence similarity 98, member B |
| chr19_-_11096677 | 0.02 |
ENSDART00000010997
|
tpm3
|
tropomyosin 3 |
| chr14_-_2179471 | 0.02 |
ENSDART00000157401
|
pcdh2g1
|
protocadherin 2 gamma 1 |
| chr14_-_2682064 | 0.02 |
ENSDART00000161677
|
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr4_-_885413 | 0.02 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr20_+_38298893 | 0.01 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr11_-_18281078 | 0.01 |
|
|
|
| chr21_+_10775209 | 0.01 |
|
|
|
| chr10_-_28316127 | 0.01 |
|
|
|
| chr5_+_22470177 | 0.01 |
ENSDART00000020434
|
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr24_+_39237314 | 0.01 |
ENSDART00000155346
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr19_-_11097075 | 0.01 |
ENSDART00000010997
|
tpm3
|
tropomyosin 3 |
| chr11_-_7020006 | 0.01 |
ENSDART00000077406
|
cdh27
|
cadherin 27 |
| chr22_-_469154 | 0.01 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr6_+_28886828 | 0.00 |
ENSDART00000130799
|
tp63
|
tumor protein p63 |
| chr10_+_1610794 | 0.00 |
ENSDART00000060946
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr13_+_11912981 | 0.00 |
ENSDART00000158244
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr12_-_41124276 | 0.00 |
ENSDART00000172022
ENSDART00000158605 ENSDART00000162967 |
dpysl4
|
dihydropyrimidinase-like 4 |
| chr1_+_23778514 | 0.00 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr13_+_11913290 | 0.00 |
ENSDART00000079398
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr9_-_6949104 | 0.00 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr24_+_39405332 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.7 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.8 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.2 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.0 | 0.3 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.1 | GO:0033634 | regulation of cell adhesion mediated by integrin(GO:0033628) positive regulation of cell adhesion mediated by integrin(GO:0033630) cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 0.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |