Project
DANIO-CODE
Navigation
Downloads
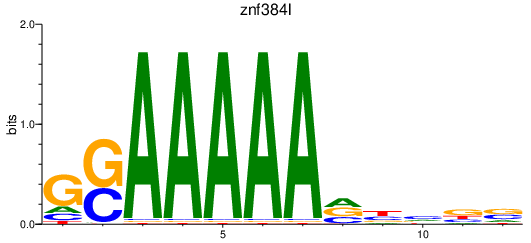
Results for znf384l
Z-value: 0.80
Transcription factors associated with znf384l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf384l
|
ENSDARG00000001015 | zinc finger protein 384 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf384l | dr10_dc_chr19_-_7069920_7069958 | 0.68 | 3.6e-03 | Click! |
Activity profile of znf384l motif
Sorted Z-values of znf384l motif
Network of associatons between targets according to the STRING database.
First level regulatory network of znf384l
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_11765407 | 1.41 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr15_+_19902697 | 1.32 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr6_+_29226868 | 1.21 |
ENSDART00000006386
|
atp1b1a
|
ATPase, Na+/K+ transporting, beta 1a polypeptide |
| chr5_+_26832823 | 1.18 |
ENSDART00000124705
|
histh1l
|
histone H1 like |
| chr7_-_26165200 | 1.16 |
ENSDART00000123395
|
her8a
|
hairy-related 8a |
| chr16_+_24063849 | 1.16 |
ENSDART00000135084
|
apoa2
|
apolipoprotein A-II |
| chr13_+_1025144 | 1.10 |
ENSDART00000148356
|
perp
|
PERP, TP53 apoptosis effector |
| chr13_+_22545318 | 1.10 |
ENSDART00000143312
|
zgc:193505
|
zgc:193505 |
| chr3_+_57878398 | 1.09 |
ENSDART00000047418
|
notum1a
|
notum pectinacetylesterase homolog 1a (Drosophila) |
| chr23_+_17939650 | 1.08 |
ENSDART00000162822
|
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr10_-_43924675 | 1.07 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr22_+_14068525 | 1.04 |
ENSDART00000080329
ENSDART00000115383 |
he1a
|
hatching enzyme 1a |
| chr15_-_35034668 | 1.02 |
ENSDART00000153787
|
rnf183
|
ring finger protein 183 |
| chr5_-_66024357 | 1.02 |
|
|
|
| chr2_-_43998886 | 0.97 |
ENSDART00000146493
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr7_+_22386456 | 0.96 |
ENSDART00000146081
|
ponzr5
|
plac8 onzin related protein 5 |
| chr3_+_51182697 | 0.95 |
|
|
|
| chr22_+_30234718 | 0.94 |
ENSDART00000172496
|
add3a
|
adducin 3 (gamma) a |
| chr3_+_24067387 | 0.94 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr3_-_25886553 | 0.93 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr7_-_2239498 | 0.93 |
ENSDART00000173892
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr18_+_5899355 | 0.92 |
|
|
|
| chr25_-_7635824 | 0.92 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr7_+_5372830 | 0.89 |
ENSDART00000135271
|
MYADM
|
myeloid associated differentiation marker |
| chr14_-_10311891 | 0.89 |
ENSDART00000136649
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr8_-_32795895 | 0.86 |
ENSDART00000131597
|
zgc:194839
|
zgc:194839 |
| chr9_-_30560440 | 0.86 |
ENSDART00000079068
|
asb11
|
ankyrin repeat and SOCS box containing 11 |
| chr5_-_39910235 | 0.85 |
ENSDART00000146237
ENSDART00000163302 |
fsta
|
follistatin a |
| chr7_+_25758469 | 0.82 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr12_-_17795448 | 0.80 |
|
|
|
| chr8_+_2428689 | 0.79 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr11_-_15162109 | 0.76 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr4_-_73485204 | 0.75 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr9_+_17431036 | 0.75 |
ENSDART00000140852
|
rgcc
|
regulator of cell cycle |
| chr23_-_18972097 | 0.74 |
ENSDART00000133826
|
CR847953.1
|
ENSDARG00000057403 |
| chr3_-_60929921 | 0.74 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr25_+_31457309 | 0.72 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr7_-_15582208 | 0.72 |
ENSDART00000102363
|
rcn1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr21_+_25651712 | 0.72 |
ENSDART00000135123
|
si:dkey-17e16.17
|
si:dkey-17e16.17 |
| chr16_+_4288700 | 0.72 |
ENSDART00000159694
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr3_-_39346621 | 0.71 |
ENSDART00000135192
ENSDART00000013553 ENSDART00000167289 |
zgc:100868
|
zgc:100868 |
| chr23_-_2084541 | 0.70 |
ENSDART00000091532
|
ndnf
|
neuron-derived neurotrophic factor |
| chr20_-_34389652 | 0.70 |
ENSDART00000144705
ENSDART00000020389 |
hmcn1
|
hemicentin 1 |
| chr3_-_23444371 | 0.69 |
ENSDART00000087726
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| KN150589v1_-_6303 | 0.69 |
|
|
|
| chr15_+_30442890 | 0.69 |
ENSDART00000048847
|
nos2b
|
nitric oxide synthase 2b, inducible |
| chr17_-_48722470 | 0.69 |
ENSDART00000160893
|
kif6
|
kinesin family member 6 |
| chr14_-_10311677 | 0.68 |
ENSDART00000133723
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr18_+_38773971 | 0.68 |
ENSDART00000010177
|
onecut1
|
one cut homeobox 1 |
| chr20_+_30948175 | 0.67 |
|
|
|
| chr25_-_3521512 | 0.67 |
ENSDART00000171863
ENSDART00000166363 |
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr3_-_54414513 | 0.66 |
ENSDART00000053106
|
s1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr12_+_30245620 | 0.66 |
ENSDART00000153116
ENSDART00000152900 |
ENSDARG00000020224
|
ENSDARG00000020224 |
| chr12_-_3742062 | 0.66 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr2_-_21265803 | 0.66 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr21_-_7480082 | 0.65 |
ENSDART00000082790
|
si:dkey-163m14.2
|
si:dkey-163m14.2 |
| chr5_-_43686611 | 0.65 |
ENSDART00000146080
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr1_-_48853800 | 0.65 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr5_-_4091643 | 0.64 |
ENSDART00000067594
|
cst14b.1
|
cystatin 14b, tandem duplicate 1 |
| chr23_+_23730717 | 0.63 |
|
|
|
| chr16_-_24697750 | 0.63 |
ENSDART00000163305
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr21_+_5693372 | 0.63 |
ENSDART00000020603
|
ccng2
|
cyclin G2 |
| chr16_+_41862759 | 0.63 |
|
|
|
| chr24_-_12794672 | 0.62 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr1_-_48790379 | 0.61 |
ENSDART00000150842
|
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr8_+_31426328 | 0.61 |
ENSDART00000135101
|
sepp1a
|
selenoprotein P, plasma, 1a |
| chr20_+_51388214 | 0.61 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr4_-_52850069 | 0.61 |
ENSDART00000169714
|
CR388413.6
|
ENSDARG00000108204 |
| chr7_+_51550533 | 0.60 |
ENSDART00000174097
|
BX957362.1
|
ENSDARG00000105682 |
| chr21_-_20674965 | 0.60 |
ENSDART00000065649
|
ENSDARG00000044676
|
ENSDARG00000044676 |
| chr18_-_36956268 | 0.60 |
|
|
|
| chr17_+_49198580 | 0.60 |
ENSDART00000059255
ENSDART00000155599 |
zgc:113176
|
zgc:113176 |
| chr16_-_29342511 | 0.59 |
ENSDART00000058870
|
rhbg
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr8_-_22945616 | 0.59 |
|
|
|
| chr12_-_8780098 | 0.59 |
ENSDART00000113148
|
jmjd1cb
|
jumonji domain containing 1Cb |
| chr1_-_46047321 | 0.58 |
|
|
|
| chr15_+_46141934 | 0.58 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr3_+_12432686 | 0.58 |
|
|
|
| chr16_+_42993841 | 0.57 |
ENSDART00000156767
|
CR854827.4
|
ENSDARG00000097750 |
| chr2_-_42384993 | 0.57 |
ENSDART00000141358
|
apom
|
apolipoprotein M |
| chr4_-_26262761 | 0.57 |
ENSDART00000176623
|
BX957270.2
|
ENSDARG00000107325 |
| chr6_+_29226824 | 0.57 |
ENSDART00000006386
|
atp1b1a
|
ATPase, Na+/K+ transporting, beta 1a polypeptide |
| chr15_-_6979997 | 0.56 |
ENSDART00000169944
|
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr13_+_29640492 | 0.56 |
ENSDART00000160944
|
pax2a
|
paired box 2a |
| chr24_-_20513975 | 0.56 |
ENSDART00000127923
|
nktr
|
natural killer cell triggering receptor |
| chr20_+_51388313 | 0.56 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr13_+_11913290 | 0.55 |
ENSDART00000079398
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_+_4557485 | 0.55 |
ENSDART00000052423
|
spry2
|
sprouty RTK signaling antagonist 2 |
| chr19_-_28699796 | 0.55 |
ENSDART00000079104
|
ndufs6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr12_-_1442400 | 0.55 |
|
|
|
| chr11_-_30105047 | 0.55 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr12_+_40919018 | 0.54 |
ENSDART00000176047
|
BX072562.1
|
ENSDARG00000108020 |
| chr13_+_48324283 | 0.54 |
ENSDART00000177067
|
CU929120.1
|
ENSDARG00000107321 |
| chr2_+_20748431 | 0.54 |
ENSDART00000137848
|
palmda
|
palmdelphin a |
| chr19_-_44043682 | 0.54 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr3_+_19186494 | 0.53 |
ENSDART00000111528
|
kri1
|
KRI1 homolog |
| chr3_+_23090476 | 0.53 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr4_-_46819684 | 0.53 |
|
|
|
| chr3_-_49242655 | 0.53 |
ENSDART00000142200
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr24_+_26287471 | 0.53 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr18_-_40893823 | 0.52 |
ENSDART00000059194
|
snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr5_-_62821458 | 0.52 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr23_+_21610798 | 0.52 |
ENSDART00000111966
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr3_+_54876680 | 0.52 |
ENSDART00000111585
|
hbbe1.3
|
hemoglobin, beta embryonic 1.3 |
| chr21_-_2147438 | 0.52 |
ENSDART00000172276
|
zgc:163077
|
zgc:163077 |
| chr7_-_7164950 | 0.52 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr11_+_29753417 | 0.51 |
|
|
|
| chr4_+_7668939 | 0.50 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr3_+_52448081 | 0.49 |
ENSDART00000154410
|
colgalt1
|
collagen beta(1-O)galactosyltransferase 1 |
| chr16_+_53178713 | 0.49 |
ENSDART00000174634
|
CABZ01062546.1
|
ENSDARG00000108858 |
| chr18_+_45203251 | 0.49 |
ENSDART00000015786
|
gyltl1b
|
glycosyltransferase-like 1b |
| chr23_+_36002332 | 0.49 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr4_+_30499948 | 0.49 |
ENSDART00000164555
|
si:dkey-199m13.4
|
si:dkey-199m13.4 |
| chr7_+_23811321 | 0.49 |
|
|
|
| chr15_-_40124634 | 0.49 |
ENSDART00000063789
|
rps5
|
ribosomal protein S5 |
| chr5_-_40451061 | 0.49 |
ENSDART00000083515
|
pdzd2
|
PDZ domain containing 2 |
| chr7_+_25649559 | 0.48 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr16_+_9871347 | 0.48 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr6_+_1873957 | 0.48 |
|
|
|
| chr15_-_35394535 | 0.48 |
ENSDART00000144153
|
mff
|
mitochondrial fission factor |
| chr4_-_52610458 | 0.48 |
|
|
|
| chr4_+_29842350 | 0.48 |
|
|
|
| chr5_-_6004819 | 0.47 |
ENSDART00000099570
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr15_+_666833 | 0.47 |
ENSDART00000157207
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr19_-_11239650 | 0.47 |
ENSDART00000146557
|
si:dkey-240h12.3
|
si:dkey-240h12.3 |
| chr13_+_44720106 | 0.47 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr13_+_11912981 | 0.46 |
ENSDART00000158244
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr20_+_43751193 | 0.46 |
ENSDART00000017269
|
parp1
|
poly (ADP-ribose) polymerase 1 |
| chr4_+_50923792 | 0.45 |
ENSDART00000161370
|
BX640547.3
|
ENSDARG00000100076 |
| chr6_+_19485784 | 0.45 |
|
|
|
| chr17_-_50343403 | 0.45 |
ENSDART00000140121
|
si:ch211-235i11.5
|
si:ch211-235i11.5 |
| chr11_+_18862603 | 0.45 |
|
|
|
| chr4_+_28860191 | 0.45 |
ENSDART00000110358
ENSDART00000162069 |
znf1059
|
zinc finger protein 1059 |
| chr19_+_43908885 | 0.44 |
ENSDART00000159421
|
BX649384.3
|
ENSDARG00000100839 |
| chr19_-_5429428 | 0.44 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr16_+_51316772 | 0.44 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr12_-_1442655 | 0.44 |
|
|
|
| chr3_-_32464649 | 0.44 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr14_+_30568242 | 0.44 |
|
|
|
| chr2_-_21677086 | 0.44 |
ENSDART00000057022
|
klhl40a
|
kelch-like family member 40a |
| chr16_-_24696036 | 0.44 |
|
|
|
| chr3_+_19186724 | 0.44 |
ENSDART00000110548
|
kri1
|
KRI1 homolog |
| chr11_+_29753292 | 0.44 |
|
|
|
| chr5_-_15774816 | 0.44 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr5_+_32231368 | 0.44 |
ENSDART00000169358
|
ENSDARG00000095142
|
ENSDARG00000095142 |
| chr23_+_36007936 | 0.44 |
ENSDART00000128533
|
hoxc3a
|
homeo box C3a |
| chr15_-_9055873 | 0.44 |
ENSDART00000124998
|
rtn2a
|
reticulon 2a |
| chr24_+_4946542 | 0.43 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| KN149973v1_+_28300 | 0.43 |
|
|
|
| chr16_+_12740993 | 0.43 |
ENSDART00000128497
|
nat14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr1_-_53746151 | 0.43 |
ENSDART00000179565
|
CABZ01043826.1
|
ENSDARG00000108130 |
| chr10_+_35321228 | 0.43 |
ENSDART00000048831
|
tmem120a
|
transmembrane protein 120A |
| chr10_-_22967786 | 0.42 |
|
|
|
| chr16_-_31042576 | 0.42 |
ENSDART00000077185
|
dgat1b
|
diacylglycerol O-acyltransferase 1b |
| chr6_-_49217110 | 0.42 |
ENSDART00000143252
ENSDART00000103376 ENSDART00000132347 ENSDART00000132131 |
ngfb
|
nerve growth factor b (beta polypeptide) |
| chr14_+_23917724 | 0.42 |
ENSDART00000138082
ENSDART00000079215 |
stc2a
|
stanniocalcin 2a |
| chr9_-_33518818 | 0.42 |
ENSDART00000140039
|
rpl8
|
ribosomal protein L8 |
| chr10_+_8670564 | 0.42 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr2_+_38178934 | 0.42 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr5_+_58017820 | 0.41 |
ENSDART00000083015
|
ccdc84
|
coiled-coil domain containing 84 |
| chr21_+_22808694 | 0.41 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr4_+_29855479 | 0.41 |
|
|
|
| chr1_+_1541977 | 0.41 |
ENSDART00000048828
|
atp1a1a.4
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 4 |
| chr3_-_12432661 | 0.40 |
|
|
|
| chr20_+_51388660 | 0.40 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr21_+_27427785 | 0.40 |
ENSDART00000113061
|
bbs1
|
Bardet-Biedl syndrome 1 |
| chr21_+_16987306 | 0.40 |
ENSDART00000080628
|
arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr16_-_39620777 | 0.40 |
ENSDART00000039832
|
tgfbr2b
|
transforming growth factor beta receptor 2b |
| chr7_+_15081529 | 0.40 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr5_-_65103664 | 0.40 |
ENSDART00000130888
|
notch1b
|
notch 1b |
| chr4_+_43151489 | 0.40 |
ENSDART00000166967
|
si:ch211-226o13.2
|
si:ch211-226o13.2 |
| chr24_-_25099449 | 0.40 |
ENSDART00000153798
|
hhla2b.2
|
HERV-H LTR-associating 2b, tandem duplicate 2 |
| chr2_+_35745559 | 0.39 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr4_+_7668691 | 0.39 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr12_-_1442470 | 0.39 |
|
|
|
| chr10_+_27090911 | 0.39 |
ENSDART00000089095
|
wdr74
|
WD repeat domain 74 |
| chr13_-_43116745 | 0.39 |
ENSDART00000161017
|
eif3s6ip
|
eukaryotic translation initiation factor 3, subunit 6 interacting protein |
| chr8_-_1904720 | 0.39 |
ENSDART00000081563
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr16_+_28449134 | 0.39 |
ENSDART00000059044
|
itga8
|
integrin, alpha 8 |
| chr13_+_22165699 | 0.38 |
|
|
|
| chr16_-_16274298 | 0.38 |
ENSDART00000103815
|
stmn2a
|
stathmin 2a |
| chr5_-_24806610 | 0.38 |
ENSDART00000089748
|
rorb
|
RAR-related orphan receptor B |
| chr22_+_1568413 | 0.38 |
ENSDART00000175704
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr19_-_6856033 | 0.38 |
ENSDART00000170952
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr19_+_23414927 | 0.38 |
ENSDART00000151090
|
gdf6b
|
growth differentiation factor 6b |
| chr24_-_3445036 | 0.38 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr13_+_3533517 | 0.38 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr3_-_55399331 | 0.37 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr16_+_12101666 | 0.37 |
ENSDART00000143442
|
p3h3
|
prolyl 3-hydroxylase 3 |
| chr7_-_32859341 | 0.37 |
ENSDART00000127006
ENSDART00000143768 |
mns1
|
meiosis-specific nuclear structural 1 |
| chr4_-_30124515 | 0.37 |
|
|
|
| chr6_-_15884727 | 0.37 |
ENSDART00000175827
|
CR847983.3
|
ENSDARG00000106155 |
| chr8_+_21321765 | 0.37 |
ENSDART00000131691
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr4_-_4241386 | 0.37 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr4_-_46037928 | 0.37 |
|
|
|
| chr24_-_25102437 | 0.37 |
|
|
|
| chr5_-_46382728 | 0.36 |
|
|
|
| chr6_-_43390195 | 0.36 |
ENSDART00000154434
|
frmd4ba
|
FERM domain containing 4Ba |
| chr25_-_3521317 | 0.36 |
ENSDART00000171863
ENSDART00000166363 |
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr4_+_70917419 | 0.36 |
ENSDART00000174035
|
si:cabz01054394.5
|
si:cabz01054394.5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0071635 | transforming growth factor beta activation(GO:0036363) transforming growth factor beta production(GO:0071604) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.4 | 1.8 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.4 | 1.1 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.4 | 1.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.3 | 1.0 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.3 | 2.0 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.2 | 1.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 0.7 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.2 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 0.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 0.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.8 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 0.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.7 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.2 | 1.0 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 0.8 | GO:0048903 | anterior lateral line neuromast development(GO:0048901) anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.2 | 0.5 | GO:0043393 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.2 | 0.6 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 0.8 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.6 | GO:0046327 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 0.4 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.4 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.9 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.4 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.1 | 0.6 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 1.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.4 | GO:0010939 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.1 | 0.4 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.4 | GO:0023058 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.1 | 0.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.2 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.2 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.2 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 1.1 | GO:0060324 | face development(GO:0060324) |
| 0.1 | 0.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.7 | GO:0070977 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.1 | 0.4 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.7 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.6 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.3 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.4 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 0.2 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.5 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 0.3 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.5 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of BMP from receptor via BMP binding(GO:0038098) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0035872 | peptidoglycan transport(GO:0015835) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.5 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0019856 | pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.2 | GO:0070178 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0060579 | amacrine cell differentiation(GO:0035881) ventral spinal cord interneuron fate commitment(GO:0060579) |
| 0.0 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 1.0 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.1 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.0 | 0.3 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.0 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.3 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.0 | 0.2 | GO:0030324 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0019346 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 1.5 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.5 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.4 | GO:0044259 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 0.2 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0051701 | interaction with host(GO:0051701) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.3 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 1.2 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0097345 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.0 | 0.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.0 | 0.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 1.0 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 1.8 | GO:1904949 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 0.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.7 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.5 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.0 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 1.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 1.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 1.6 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.5 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.3 | 1.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 0.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 0.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.6 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.4 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 0.5 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.4 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.4 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.7 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 1.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.4 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 2.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 1.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.5 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.9 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |