Project
DANIO-CODE
Navigation
Downloads
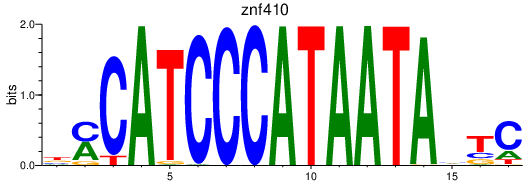
Results for znf410
Z-value: 0.57
Transcription factors associated with znf410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf410
|
ENSDARG00000008218 | zinc finger protein 410 |
Activity profile of znf410 motif
Sorted Z-values of znf410 motif
Network of associatons between targets according to the STRING database.
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_5142209 | 3.78 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr19_-_5142238 | 3.00 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr16_-_31835083 | 1.79 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr19_-_5142009 | 1.63 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr16_-_31835007 | 1.23 |
|
|
|
| chr6_+_21887935 | 0.86 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr6_+_21887963 | 0.66 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr21_-_14735450 | 0.65 |
ENSDART00000144859
|
pus1
|
pseudouridylate synthase 1 |
| chr3_+_18417755 | 0.08 |
ENSDART00000124329
|
cbx8a
|
chromobox homolog 8a (Pc class homolog, Drosophila) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1990481 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 6.3 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 1.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |